cat no | io1001
ioGlutamatergic Neurons
Human iPSC-derived glutamatergic neurons
-
Cryopreserved human iPSC-derived cells powered by opti-ox, that are ready for experiments in days
-
Ideal for studying excitatory signalling pathways, neurodegenerative diseases and neurotoxicology
-
Consistent, functional excitatory neurons, with no inhibitory neurons

Human iPSC-derived glutamatergic neurons
ioGlutamatergic Neurons display neuronal activity that matures over time
The function of ioGlutamatergic Neurons was investigated using the MaxTwo HD-MEA System.
The Axon Tracking Assay (left) shows examples of reconstructed axonal paths of travelling action potentials of individual iPSC-derived glutamatergic neurons. The assay reveals the spatial propagation of the neuronal action potential from the soma to distant axonal branches.
Total axon length (middle) and firing rate (right) increase over time, indicating that the cells are maturing. ioGlutamatergic Neurons were cultured with human iPSC-derived astrocytes.
Data courtesy of Charles River Laboratories and MaxWell Biosystems.

Rapid maturation of ioGlutamatergic Neurons leads to synchronised network activity by day 31
Raster plots generated using the MaxTwo HD-MEA System show the development of the neuronal network over time.
The plots show the dynamics of the network activity using 1,024 active electrodes. Each row represents an individual electrode and each blue dot indicates a spike detected at that electrode over a period of 300 seconds.
Spontaneous activity is observed at DIV 7. Clear synchronised bursting activity is observed by DIV 31, represented by blue vertical lines, followed by an overall drop in activity, seen as white lines. ioGlutamatergic Neurons were cultured with human iPSC-derived astrocytes.
Download our poster to see additional data that shows how ioGABAergic Neurons form functional neuronal networks with ioGlutamatergic Neurons in the presence of astrocytes, and how the tri-culture responds to bicuculline and diazepam.
ioGlutamatergic Neurons offer a rapidly maturing functional system that can be used to assess neuronal networks and the impact of a drug treatment or intervention.
Data courtesy of Charles River Laboratories and MaxWell Biosystems.

ioGlutamatergic Neurons offer a robust, physiologically-relevant model for efficacy screening of candidate ASOs
Positive and negative control antisense oligonucleotides (ASOs) with gapmer chemistry were introduced into glutamatergic neurons by gymnosis. RT-qPCR was used to measure ASO-induced gene knockdown.
- Strong separation of the assay signal for positive control (blue) and negative control (orange) ASOs was observed for all plates tested (A).
- The positive control ASO induced ~90% knockdown of the target gene, shown by a decrease in the target gene expression (A) and higher Cp (or Ct) values for the target gene, indicating lower initial amount of the target sequence (B).
- There was no effect of the control ASOs on housekeeping gene expression as compared to vehicle-transfected controls (C).
- No marked intra- or inter-plate variability was observed between positive and negative control ASOs (A-C).
Data courtesy of Charles River Laboratories.

ioGlutamatergic Neurons show good suitability for high-throughput screening in 384-well format plates
Cytotoxicity CellTiter-Glo®️ (CTG) and TR-FRET (HTRF®️) assays for AKT serine/threonine kinase 1 (AKT) and Huntingtin (HTT) proteins were performed on ioGlutamatergic Neurons in 384-well plates treated with tool compound (cmp) at day 9 post-revival. Compound titration results in a concentration response curve for all three assays (mean±sd of 2 replicates). CTG assay on ioGlutamatergic Neurons shows an excellent average signal-to-background ratio and high suitability for HTS. HTRF assays on ioGlutamatergic Neurons show lower signals but with low variability, and could therefore also provide a suitable platform for HTS.
Data courtesy of Charles River Laboratories.

Whole transcriptome analysis demonstrates high lot-to-lot consistency across three manufactured lots of ioGlutamatergic Neurons
Bulk RNA-sequencing analysis was performed on three different lots of ioGlutamatergic Neurons on day 0, day 11 and day 18 post-revival. (A) A principal component analysis (PCA) to assess gene expression variance between three different manufactured lots showed a tight clustering of the samples at each timepoint, demonstrating high consistency between these lots. This lot-to-lot consistency of ioGlutamatergic Neurons will help reduce experimental variation and increase the reproducibility of experiments. (B) PCA without the parental non-induced hiPSC samples, highlighting the tight clustering of the day 11 as well as day 18 samples of the three different lots. (C) Differential expression test reveals no statistically significant differentially expressed (DE) genes across the three lots at day 11 (|logFC| > 0.5 and FDR < 0.01).
Colours represent the three lots of products; shapes represent the parental non-induced hiPSC line and different timepoints.
Expression levels for specific genes of interest can be requested by contacting our team at technical@bit.bio.

High lot-to-lot consistency is demonstrated by a consistent transcriptomic fingerprint across manufactured lots of ioGlutamatergic Neurons
Single cell RNA-sequencing analysis was performed on three different lots of ioGlutamatergic Neurons on day 11. UMAP plots represent the cell-to-cell variation in gene expression profiles of cells, each dot representing an individual cell. Cells from each of the three lots are equally distributed across the body of the plot. Merging the UMAP plots creates a tight overlay, showing a strong transcriptional relationship between cells from three independently manufactured lots of ioGlutamatergic Neurons. Gene expression was assessed by 10x Genomics scRNA-sequencing.

Single cell ATAC-sequencing shows a consistent transcriptomic fingerprint demonstrating high lot-to-lot consistency across manufactured lots of ioGlutamatergic Neurons
Single cell ATAC-sequencing analysis was performed on three different lots of ioGlutamatergic Neurons on day 11. Single cell ATAC-sequencing reveals regions of open chromatin to understand the gene regulatory landscape of individual cells. UMAP plots represent the cell-to-cell variation in chromatin accessibility of the cells, each dot representing a single cell. Cells from each of the three lots are equally distributed across the body of the plot. Merging the UMAP plots creates a tight overlay, showing a strong transcriptional relationship between cells from three independently manufactured lots of ioGlutamatergic Neurons. Gene expression was assessed by 10x Genomics scRNA-sequencing.
ioGlutamatergic Neurons generated by transcription factor-driven deterministic cell programming of iPSCs using opti-ox technology
Time-lapse video capturing the rapid and homogeneous neuronal phenotype acquisition upon thawing of cryopreserved ioGlutamatergic Neurons. 7 day time course.

ioMicroglia enhance network activity in co-culture with ioGlutamatergic Neurons
ioGlutamatergic Neurons expressing Incucyte® Neuroburst Orange Lentivirus mono-culture or in co-culture with
ioMicroglia Male (io1021) monitored and quantified using Incucyte® Neuronal Activity Analysis software.
A) Representative calcium traces shown for each culture condition at 15 days post-microglia addition.
B) Bar charts at 15 days post-microglia addition showing network correlation and mean burst duration. Data presented as mean ± SEM, n = 3 – 12 replicates.
This data was generated by Jasmine Trigg and colleagues at Sartorius, taken from the application note: "Advanced in vitro Modeling of Human iPSC-derived Neuronal Mono- and Co-cultures with Microglia: Optimization Using Growth Factors and Live-Cell Analysis".

ioGABAergic Neurons exert an inhibitory effect on the excitatory ioGlutamatergic Neurons within the tri-cultures leading to a higher network burst rate
The effect of adding increasing numbers of inhibitory ioGABAergic Neurons (io1003) to the tri-cultures was investigated by MEA analysis at 53 DIV, alongside the control co-cultures. Representative raster plots displaying the activity of 16 electrodes over a time period of 300 seconds are shown. Each horizontal row of the raster plot represents the activity of an electrode, within which each vertical black dash indicates a firing event, a blue dash indicates a single electrode burst, and a pink box indicates a network burst event. The histogram trace on top of the raster plot is a measure of the number of spikes per network burst. The co-culture with ioGlutamatergic Neurons and astrocytes shows the strongest network bursts as indicated by the increased number of spikes per network burst and shows a lower network burst rate (NBR) compared to the tri-cultures. The addition of increasing numbers of inhibitory ioGABAergic Neurons to the tri-cultures reduces the number of spikes per network burst and leads to an increased NBR. This indicates that ioGABAergic Neurons are having an inhibitory effect on the excitatory ioGlutamatergic Neurons. The co-culture of ioGABAergic Neurons and astrocytes shows no network bursts, indicating the absence of excitatory neurons and that the population of ioGABAergic Neurons is highly pure. Analysis was performed on an Axion Maestro Pro MEA platform. This data was generated in partnership with Charles River Laboratories.

Long-term maintenance and myelination marker expression in ioGlutamatergic Neurons and ioOligodendrocyte-like cells co-culture
ioGlutamatergic Neurons and ioOligodendrocyte-like cells were co-cultured in media containing 1 µM or 100 µM db-cAMP and fixed at timepoints day 14 and 21. Immunocytochemistry was performed for MBP (green) and TUBB3 (red). Nuclei were counterstained with DAPI (blue). Images were acquired using a Zeiss LSM 900 confocal microscope (scale bar: 50 µm).
Representative confocal images illustrate potential neuronal–oligodendrocyte interactions and sustained myelination marker expression.
View the complete dataset and the step-by-step co-culture protocol

Lipid-based delivery of synthetic mRNA into ioGlutamatergic Neurons
ioGlutamatergic Neurons were transfected 24 hours post-thaw using Lipofectamine™ Stem Transfection Reagent. The transfection efficiency was evaluated by fluorescence imaging over 18 days after mRNA delivery, resulting in high transfection efficiency (close to 100%) and long-term sustained GFP expression.
Quantification of the GFP signal shows a decrease in GFP intensity over time, while the percentage of GFP-positive cells remains largely unchanged over time.
(A) The percentage of GFP-positive cells from two independent experiments.
(B) GFP intensity, quantified in successfully transfected cells from two independent experiments is quantified and normalised to day 2 (24 hours post-transfection).

Increased ratio of A𝛽42:40 seen in ioGlutamatergic Neurons APP V717I (London), as observed in Alzheimer’s disease, vs wild-type control
Wild type ioGlutamatergic Neurons can be paired with engineered disease model cells to investigate the impact of disease-related mutations, for example, the APP missense mutation, investigating its role in early-onset Alzheimer's disease.
ioGlutamatergic Neurons APP V717I
disease model cells show increased production of A𝛽38 and A𝛽42 peptides (involved in the amyloidogenic pathway), with no difference seen for A𝛽40 (A). This results in an increased ratio of A𝛽42:40 and no change in the A𝛽42:38 ratio (B).
- ioGlutamatergic Neurons wild type (WT), APP V717I/WT (CL35, io1067S), and APP V717I/V717I (CL27, io1063S), were seeded at 30,000 cells/cm2 in 24-well plates and cultured for 30 days according to the user manual. Supernatant was collected at days 10, 20, and 30.
- Levels of A𝛽38, A𝛽40 and A𝛽42 peptides were quantified using the V-PLEX A𝛽 Peptide Panel 1 (6E10) Kit (MSD K15200E-1).
- Concentrations of A𝛽38, A𝛽40, A𝛽42 were normalised to the calculated total number of cells per well.
- Data were obtained from two independent experiments and are shown as mean ± SEM. Data were analysed statistically (at days 20 and 30) using Student’s t-tests comparing each clone to the wild type.
* p<0.05 ** p<0.01 ***p<0.001

Live-cell imaging reveals clear visualisation of GFP ioMicroglia when co-cultured with ioGlutamatergic Neurons
ioGlutamatergic Neurons were cultured to day 10 post-thaw. GFP ioMicroglia (io1096) were cultured to day 10 post-thaw and were directly added to day 11 ioGlutamatergic Neurons. The co-cultures were maintained for a further 3 days before live-cell imaging with Leica DMi8. Brightfield and fluorescence images were taken and merged, easily demonstrating distribution of GFP ioMicroglia within the co-culture.

A pooled knockout screen of neurodegenerative disease-relevant genes in CRISPRko-Ready ioGlutamatergic Neurons shows clustering of aaRS genes in UMAPs
ioGlutamatergic Neurons have been engineered to constitutively express Cas9 nuclease - CRISPRko-Ready ioGlutamatergic Neurons (io1090).
For a pooled knockout screen in CRISPRko-Ready ioGlutamatergic Neurons, 100 known genes involved in neurodegenerative diseases were selected. Lentiviral transduction of the gRNAs was carried out on day 3 and single-cell gene expression analysis was performed on day 12. Single cells were clustered on uniform manifold approximations and projections (UMAPs) based on their shared nearest neighbour’s gene expression. Clustering of aminoacyl-tRNA synthetase (aaRS) knockouts including AARS1, HARS1, CARS1, and GARS1 was observed. In contrast, cells transduced with non-targeting control sgRNAs were evenly distributed among clusters. Pathway analysis showed gRNAs targeting aaRSs activated the unfolded protein response (UPR), the mechanism by which cells control endoplasmic reticulum protein homeostasis. In many neurodegenerative diseases, signs of UPR activation have been reported. The most common aaRS-associated monogenic disorder is the incurable neurodegenerative disease Charcot–Marie–Tooth neuropathy (CMT).

Efficient mRNA transfection into ioGlutamatergic Neurons
ioGlutamatergic Neurons are efficiently transfected and show sustained long-term expression of mRNA encoding GFP. ioGlutamatergic Neurons were imaged from day 1 post-thaw and throughout the experiment to assess transfection efficiency and evaluate potential cytotoxic effects of the transfection protocol. Day 1 images were captured prior to transfection on the same day.
Download the step-by-step protocol for lipid-based delivery of synthetic mRNA into ioGlutamatergic Neurons.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.






.webp?width=2216&height=2216&name=bit.bio%20ioGlutamatergic%20Neurons%20-%20Day%2011%20-%20MAP2%20(1).webp)


%20MAP2(r)%20DAPI(b)%20(1).webp?width=2216&height=2216&name=bit.bio%20ioGlutamatergic%20Neurons%20-%20Day%2011%20-%20MERGE%20VGLUT2(g)%20MAP2(r)%20DAPI(b)%20(1).webp)








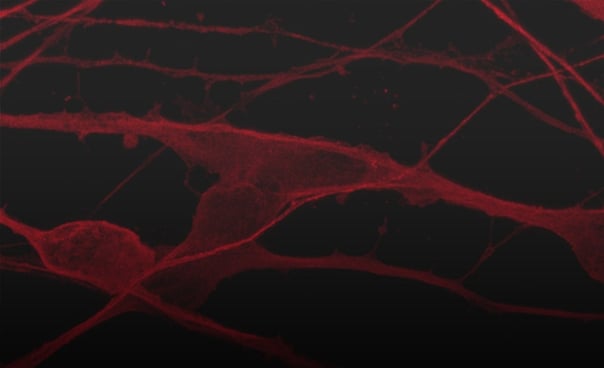
Hoescht(blue)_day12v2.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_20xMAP2(red)Hoescht(blue)_day12v2.png)
Hoescht(blue)TUBB3(blue)_day4.jpg?width=604&name=bit.bio_ioGlutamatergic%20Neurons_60xMAP2(red)Hoescht(blue)TUBB3(blue)_day4.jpg)
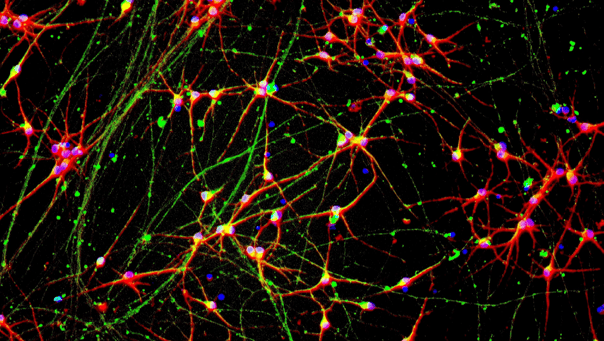

.png?width=1860&height=1260&name=bit.bio_3x2_ioGlutamatergic%20Neurons_MAP2_Hoescht_x20_hi.res%20(1).png)