













cat no | ioEA1005
ioGlutamatergic Neurons TDP‑43 M337V/M337V
Human iPSC-derived ALS and FTD disease model
-
Cryopreserved human iPSC-derived cells powered by opti-ox that are ready for experiments in days
-
Excitatory neurons with a disease-relevant mutation in TDP-43 for modelling ALS and FTD
-
Reduced neuronal activity phenotype in MEA compared to the wild-type control

Human iPSC-derived ALS and FTD disease model

ioGlutamatergic Neurons TDP-43 M337V/M337V show reduced neuronal activity indicating their potential as a relevant translational in vitro drug discovery model for ALS and FTD
Reduced neuronal activity was measured in ioGlutamatergic Neurons TDP-43 M337V/M337V compared to ioGlutamatergic Neurons TDP-43 M337V/WT and the genetically matched control, ioGlutamatergic Neurons.
Microelectrode array (MEA) chips were spotted with 100K (~900K cells/cm2) ioGlutamatergic Neurons (WT), TDP-43 M337V/WT, or TDP-43 M337V/M337V, along with 20K (~180K cells/cm2) human iPSC-derived astrocytes. From DIV6, neurobasal medium was switched to BrainPhys medium by performing a half-medium change every 48 hrs.
Brightfield at 26 DIV (A, left), cells show good coverage of electrodes and produce clear burst and network burst activity as seen in the raster plot of activity (A, right). In the raster plot, each dash indicates a firing event, blue indicates a single electrode burst and the pink box indicates a network burst event.
Quantification of raster plots over the course of culture shows that ioGlutamatergic Neurons TDP-43 M337V/M337V have a lower weighted mean firing rate and network burst frequency than WT and ioGlutamatergic Neurons TDP-43 M337V/WT (B). No clear difference is noted between WT and TDP-43 M337V/WT. Error bars indicate SEM, n=14 technical repeats.
Data courtesy of Charles River Laboratories.

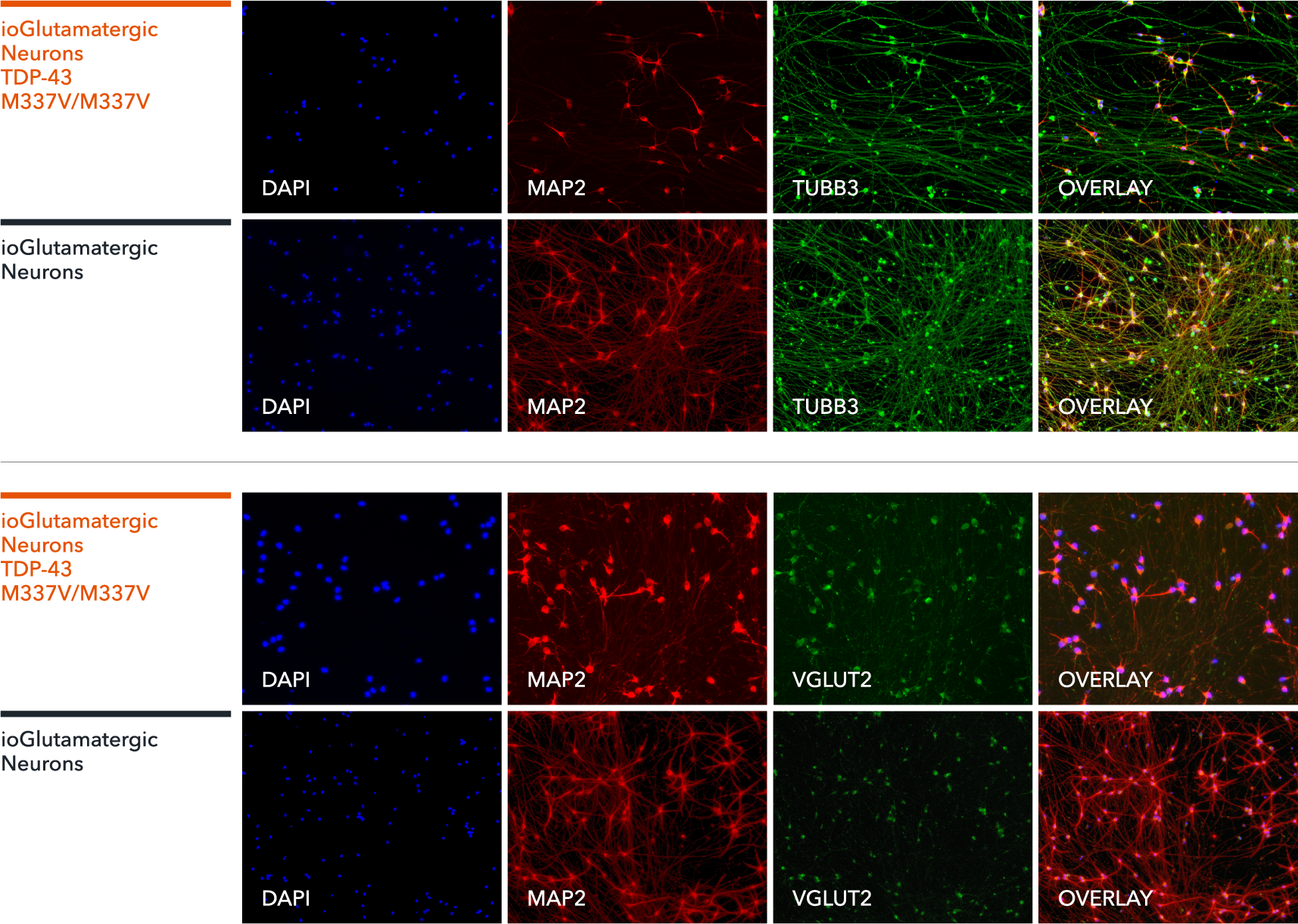
ioGlutamatergic Neurons TDP‑43 M337V/M337V express neuron-specific markers comparably to the genetically matched control
Immunofluorescent staining on day 11 post-revival demonstrates similar homogenous expression of pan-neuronal proteins MAP2 and TUBB3 (upper panel) and glutamatergic neuron-specific transporter VGLUT2 (lower panel) in ioGlutamatergic Neurons TDP‑43 M337V/M337V compared to the genetically matched control. 100X magnification.

ioGlutamatergic Neurons TDP‑43 M337V/M337V form structural neuronal networks by day 11
ioGlutamatergic Neurons TDP‑43 M337V/M337V mature rapidly, show glutamatergic neuron morphology and form structural neuronal networks over 11 days when compared to the genetically matched control. Day 1 to 11 post-thawing; 100X magnification.

ioGlutamatergic Neurons TDP‑43 M337V/M337V demonstrate gene expression of neuronal-specific and glutamatergic-specific markers following deterministic programming
Gene expression analysis demonstrates that ioGlutamatergic Neurons TDP‑43 M337V/M337V and the genetically matched, wild type control (WT) lack the expression of pluripotency markers (NANOG and OCT4), at day 11, while robustly expressing pan-neuronal (TUBB3 and SYP) and glutamatergic-specific (VGLUT1 and VGLUT2) markers, and the glutamate receptor GRIA4. Gene expression levels were assessed by RT-qPCR (data normalised to HMBS; cDNA samples of the parental human iPSC line (hiPSC Control) were included as reference). Data represents day 11 post-revival samples, n=2 replicates.

Disease-related TARDBP is expressed in ioGlutamatergic Neurons TDP‑43 M337V/M337V following deterministic programming
Gene expression analysis demonstrates that ioGlutamatergic Neurons TDP‑43 M337V/M337V and the genetically matched, wild-type control (WT) express the TARDBP gene encoding TDP‑43. Gene expression levels were assessed by RT-qPCR (data normalised to HMBS; cDNA samples of the parental human iPSC line (hiPSC Control) were included as reference). Data represents day 11 post-revival samples, n=2 replicates.

Do more with every vial
The recommended minimum seeding density is 30,000 cells/cm2, compared to up to 250,000 cells/cm2 for other similar products on the market. One small vial can plate a minimum of 0.7 x 24-well plate, 1 x 96-well plate, or 1.5 x 384-well plates. One large vial can plate a minimum of 3.6 x 24-well plates, 5.4 x 96-well plates, or 7.75 x 384-well plates. This means every vial goes further, enabling more experimental conditions and more repeats, resulting in more confidence in the data.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.










.png?width=1860&height=1260&name=bit.bio_3x2_ioGlutamatergic%20Neurons_MAP2_Hoescht_x20_hi.res%20(1).png)






