Your trusted partner in CRISPR-based functional genomic screening
of human cells
CRISPR screening has transformed the way scientists discover and validate genetic targets, helping to unravel disease mechanisms and identify new therapeutic avenues. However, traditional models often fail to capture the full complexity of human biology.
Our CRISPR-Ready ioCells powered by opti-ox provide a physiologically relevant model for performing functional genomics. Despite the potential of CRISPR screening in human iPSC-derived cells, its full power can be difficult to harness.
Let us help.
With our advanced CRISPR screening platform, you can easily outsource the design, development, and execution of high-throughput CRISPR screens, while focusing on the data. Our experts work closely with you to build and run fully customised screens tailored to your research needs.
We offer custom CRISPR screens using our existing CRISPR-Ready ioCells. Alternatively, we can collaborate with you to make any of our portfolio of human iPSC-derived cells, ioWild Type Cells or ioDisease Model Cells, ready for screening.

We currently have CRISPRko-Ready ioGlutamatergic Neurons, CRISPRko-Ready ioMicroglia, CRISPRa-Ready ioGlutamatergic Neurons and CRISPRi-Ready ioGlutamatergic Neurons, which can be used in our screening platform. If you are interested in performing a screen in a different ioWild Type Cell or ioDisease Model Cell, speak with us about generating the right model system for your screen.
Study the effect of knocking out tens to thousands of genes in one screen. Whether you want to study a particular pathway or perform a genome-wide screen, our expert team of scientists and bioinformaticians are here to design a custom sgRNA library based on your needs.
With a range of readouts including high-throughput viability assays, fluorescence-based phenotypic screens, next-generation sequencing (NGS) to identify gene knockout effects and single-cell RNA sequencing, our platform enables you to discover key genetic drivers of biological processes. We provide the tools and expertise to deliver high-resolution, data-rich insights tailored to your research needs.
Need more support? We offer full data analysis support for your CRISPR screening results.

HNRNPH1 regulates the neuroprotective cold‐shock protein RBM3 expression through poison exon exclusion
In this paper, the team of researchers used CRISPR-Cas9 to systematically knockout every gene in the human genome, one at a time in a pooled screen, in human iPSC-derived glutamatergic neurons, with the goal of identifying genes that regulate the neuroprotective cold shock protein RBM3.
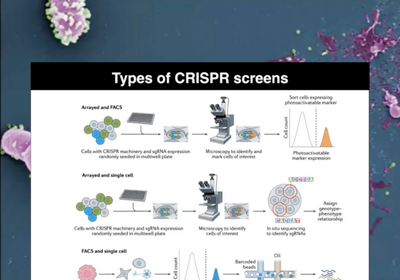
Applying human cells in CRISPR screens
This webinar explores how CRISPR-Cas9 is a vital tool for functional genomics, aiding in identifying genomic variations linked to diseases and validating drug targets. The experts also discuss scalable tools for gene characterisation and advances in leveraging human iPSC-derived cell technologies for CRISPR-based knockouts.

CRISPR knockout screening in human iPSC-derived ioMicroglia
In this scientific poster, CRISPR-Ready ioMicroglia are characterised, including cell morphology and gene expression profiles, functional Cas9 expression, single gene knockout efficiency, and applicability to pooled CRISPR screens.
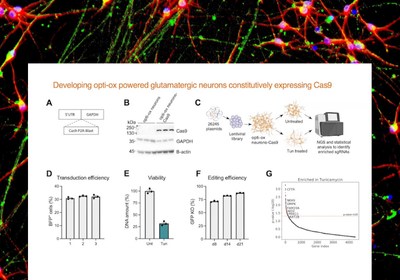
Discover drug targets for neurodegeneration
This webinar explores two peer-reviewed studies using human neurons powered by opti-ox constitutively expressing Cas9 for the generation of large scale CRISPR screens for the identification of new therapeutic targets for the treatment of neurodegenerative diseases.

CRISPR-Cas9 knockout screen in iPSC-derived Neurons identifies new Alzheimer’s disease druggable target
In this paper, human iPSC-derived neurons powered by opti-ox, constitutively expressing Cas9, enable a pooled CRISPR screen targeting the human druggable genome. KAT2B is identified as a target from the screen, which when inhibited in the presence of Tunicamycin attenuates neuronal cell death.
Using CRISPRko-Ready, CRISPRi-Ready and CRISPRa-Ready ioCells eliminates the need to spend months engineering and characterising Cas9-stable iPSC lines and optimising differentiation protocols, significantly reducing experimental timelines. With these ready-to-screen cells, reliable and reproducible experimental results can be achieved quickly.
Discover the data generated from CRISPR-Ready ioCells below.
A pooled single cell CRISPR knockout screen uncovers modulators of microglia activation.

To conduct a pooled single cell CRISPR knockout screen (scCRISPR screen) with a targeted sequencing readout, we first identified a transcriptomic activation signature of 258 differentially expressed genes by comparing CRISPR-Ready ioMicroglia treated with and without LPS at day 10. Separately, we selected 110 candidate genes for the pooled scCRISPR screen based on their known roles in neurodegeneration and neuroinflammation. Guide RNAs were delivered via lentiviral transduction on day 10, aiming for a single integration per cell. The cells were cultured and then treated with +/- LPS for 24 hours before single cell processing on day 15. Cosine similarity analysis compared knockouts in LPS-treated CRISPR-Ready ioMicroglia to both resting and activated states. The analysis identified 17 gene knockouts that altered responses to LPS stimulation. The heatmap shows Log2FC profiles for gene knockouts that had a cosine similarity above 0.3 (arbitrarily chosen threshold) compared to cells with non-targeting guides in the unstimulated condition. Knockouts are sorted based on their cosine similarity to the non-LPS condition. CD14, MAP3K7, TIRAP, IKBKG, TRAF6, IKBKB, LY96, TICAM1, RELA, and TLR4 are genes known to be involved in LPS activation mediated via the TLR4 signalling pathway.
A pooled knockout screen of neurodegenerative disease-relevant genes in CRISPR-Ready ioGlutamatergic Neurons shows clustering of aaRS genes in UMAPs

If we don't currently have the cells your target identification and validation workflows need, our team of experts can develop new CRISPR-Ready ioCells in any ioWild Type Cell background. Additionally, if you are looking to outsource the development of high throughput CRISPR screens in CRISPR-Ready ioCells, we can design, build and run these for you.
