













cat no | io1059
ioGlutamatergic Neurons APP KM670/671NL / KM670/671NL
Human iPSC-derived Alzheimer's disease model
- Cryopreserved human iPSC-derived cells powered by opti-ox that are ready for experiments in days
- Functional excitatory neurons engineered with the APP Swedish mutation for Alzheimer's disease research
- Disease-related phenotype demonstrated by increased amyloid beta production vs. wild-type control

Human iPSC-derived Alzheimer's disease model

Increased overall production of A𝛽38, A𝛽40 and A𝛽42 shown in ioGlutamatergic Neurons APP KM670/671NL (Swedish), as observed in Alzheimer’s disease
ioGlutamatergic Neurons APP KM670/671NL Alzheimer's disease model cells show an increase in the overall production of A𝛽38, A𝛽40 and A𝛽42 compared to the wild type, genetically matched control (A), and no change in the ratios of the A𝛽 peptides (B).
- ioGlutamatergic Neurons wild type (WT, io1001) and APP KM670/671NL Het (CLE4, io1061S) and Hom (CLH12, io1059S), were seeded at 30,000 cells/cm2 in 24-well plates and cultured for 30 days according to the user manual. Supernatant was collected at days 10, 20, and 30.
- Levels of A𝛽38, A𝛽40 and A𝛽42 peptides were quantified using the V-PLEX A𝛽 Peptide Panel 1 (6E10) Kit (MSD K15200E-1).
- Concentrations of A𝛽38, A𝛽40, A𝛽42 were normalised to the calculated total number of cells per well.
- Data were obtained from two independent experiments and are shown as mean ± SEM. Data were analysed statistically (at days 20 and 30) using Student’s t-tests comparing each disease model to the wild type.
* p<0.05 ** p<0.01

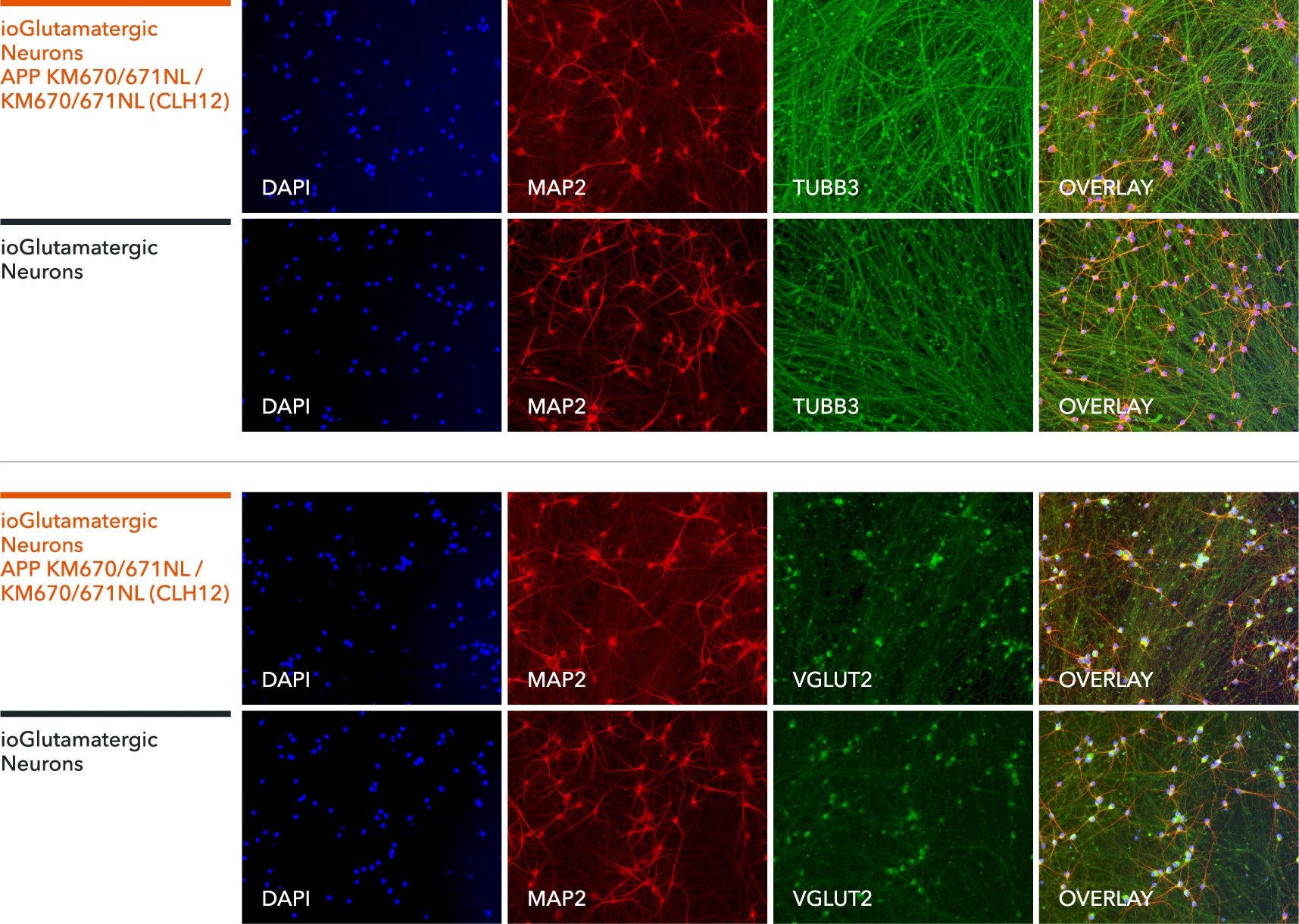
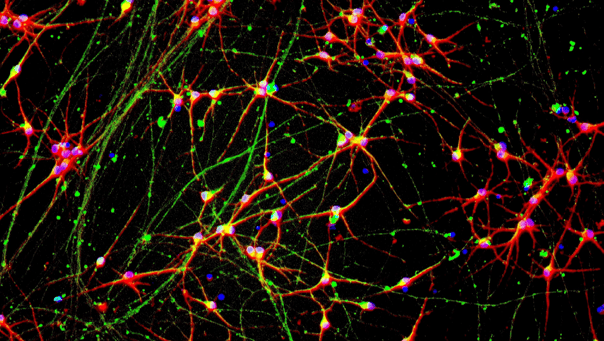
ioGlutamatergic Neurons APP KM670/671NL hom express neuron specific markers comparably to the genetically matched control
Immunofluorescent staining on post-revival day 11 demonstrates similar homogenous expression of pan-neuronal proteins TUBB3 and MAP2 (upper panel) and glutamatergic neuron specific transporter VGLUT2 (lower panel) in glutamatergic neurons carrying the Alzheimer's disease related APP KM670/671NL mutation compared to the genetically matched control. 100X magnification.

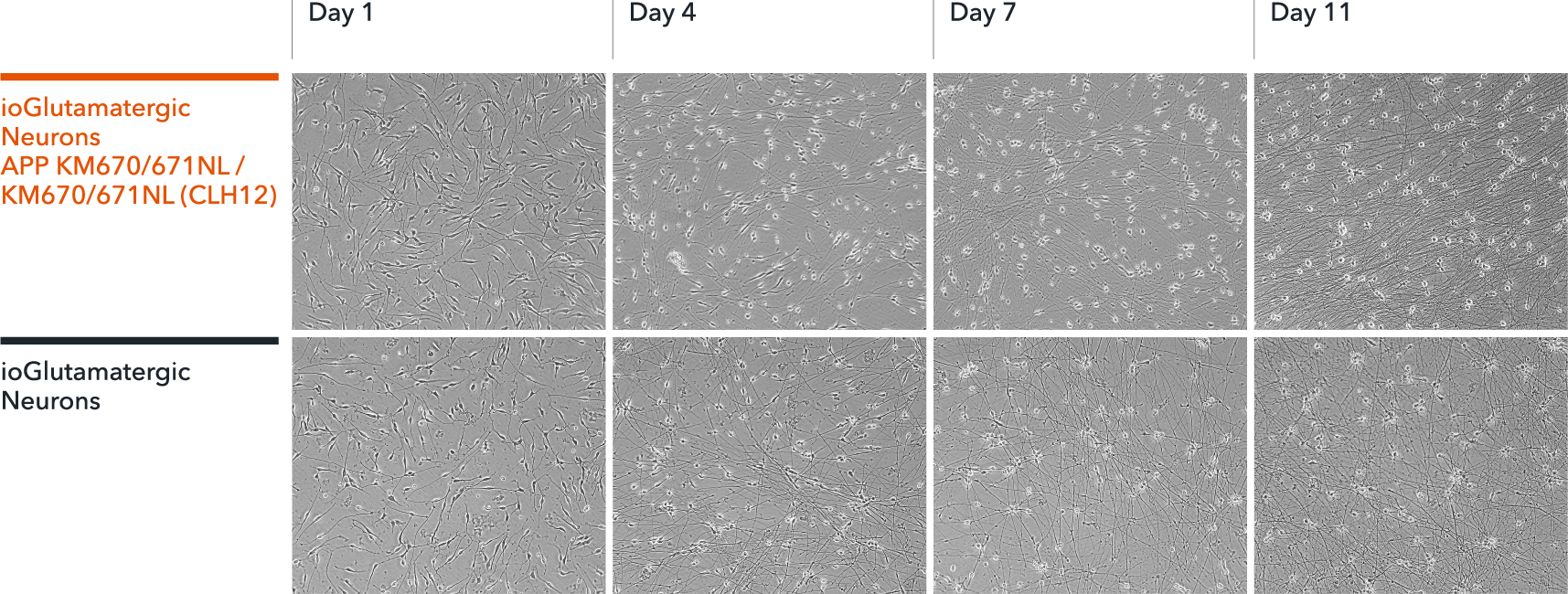
ioGlutamatergic Neurons APP KM670/671NL / KM670/671NL form structural neuronal networks by day 11
ioGlutamatergic Neurons APP KM670/671NL hom mature rapidly, show glutamatergic neuron morphology and form structural neuronal networks over 11 days, highly similar to the genetically matched control. Day 1 to 11 post thawing; 100X magnification.

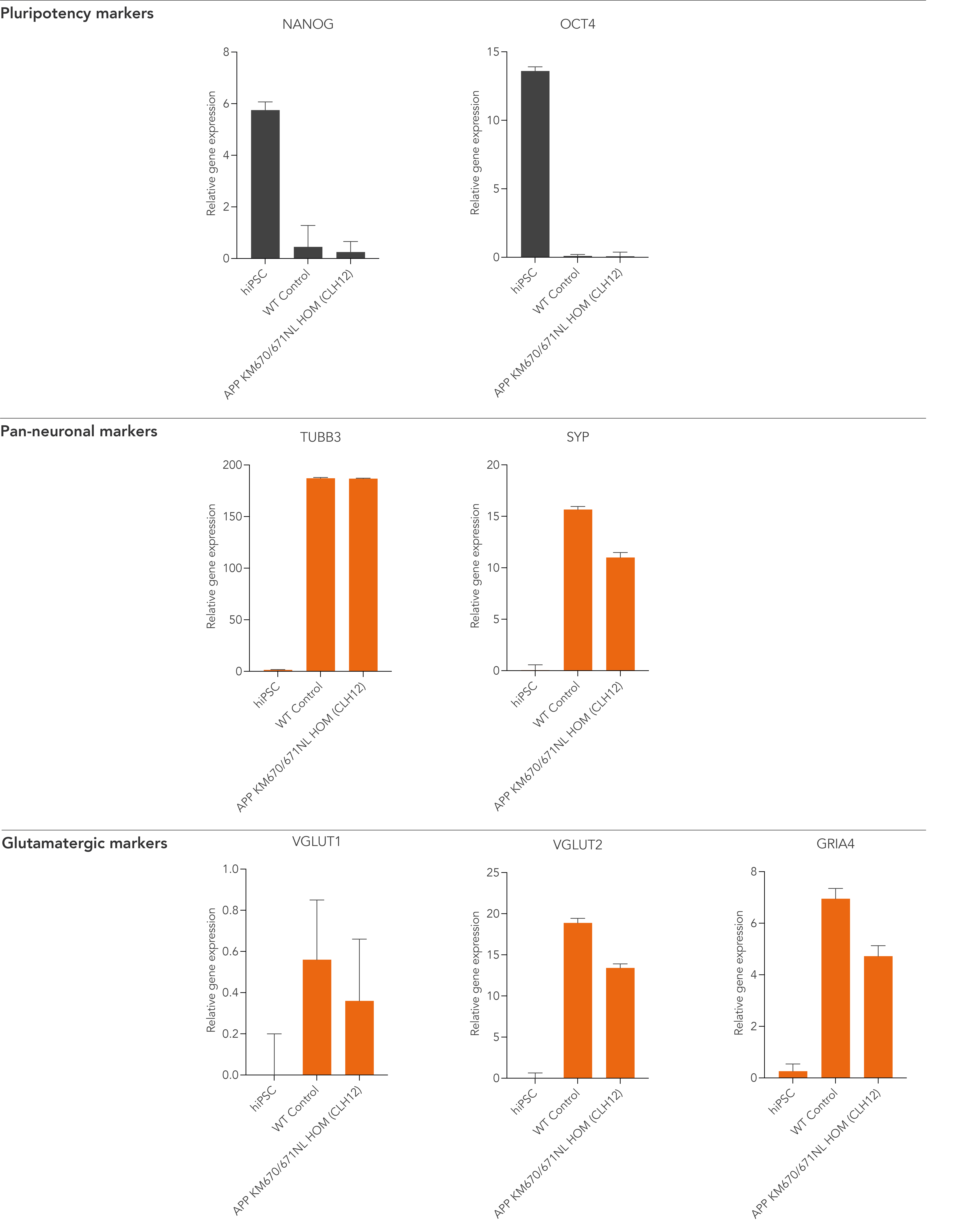
ioGlutamatergic Neurons APP KM670/671NL / KM670/671NL demonstrate gene expression of neuronal and glutamatergic-specific markers following deterministic programming
Gene expression analysis demonstrates that ioGlutamatergic Neurons APP KM670/671NL hom and the genetically matched control (WT Control) lack the expression of pluripotency markers (NANOG and OCT4) at day 11, whilst robustly expressing pan-neuronal (TUBB3 and SYP) and glutamatergic markers (VGLUT1 and VGLUT2), as well as the glutamate receptor GRIA4. Gene expression levels were assessed by RT-qPCR (data normalised to HMBS; cDNA samples of the parental human iPSC line (hiPSC) were included as reference). Data represents day 11 post-revival samples, n=2 replicates.

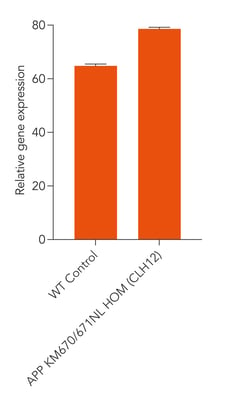
Disease-related APP is expressed in ioGlutamatergic Neurons APP KM670/671NL / KM670/671NL following deterministic programming
Gene expression analysis demonstrates that ioGlutamatergic Neurons APP KM670/671NL hom and the genetically matched control (WT Control) express the APP gene encoding the amyloid precursor protein. Gene expression levels were assessed by RT-qPCR (data normalised to HMBS). Data represents day 11 post-revival samples, n=2 replicates.

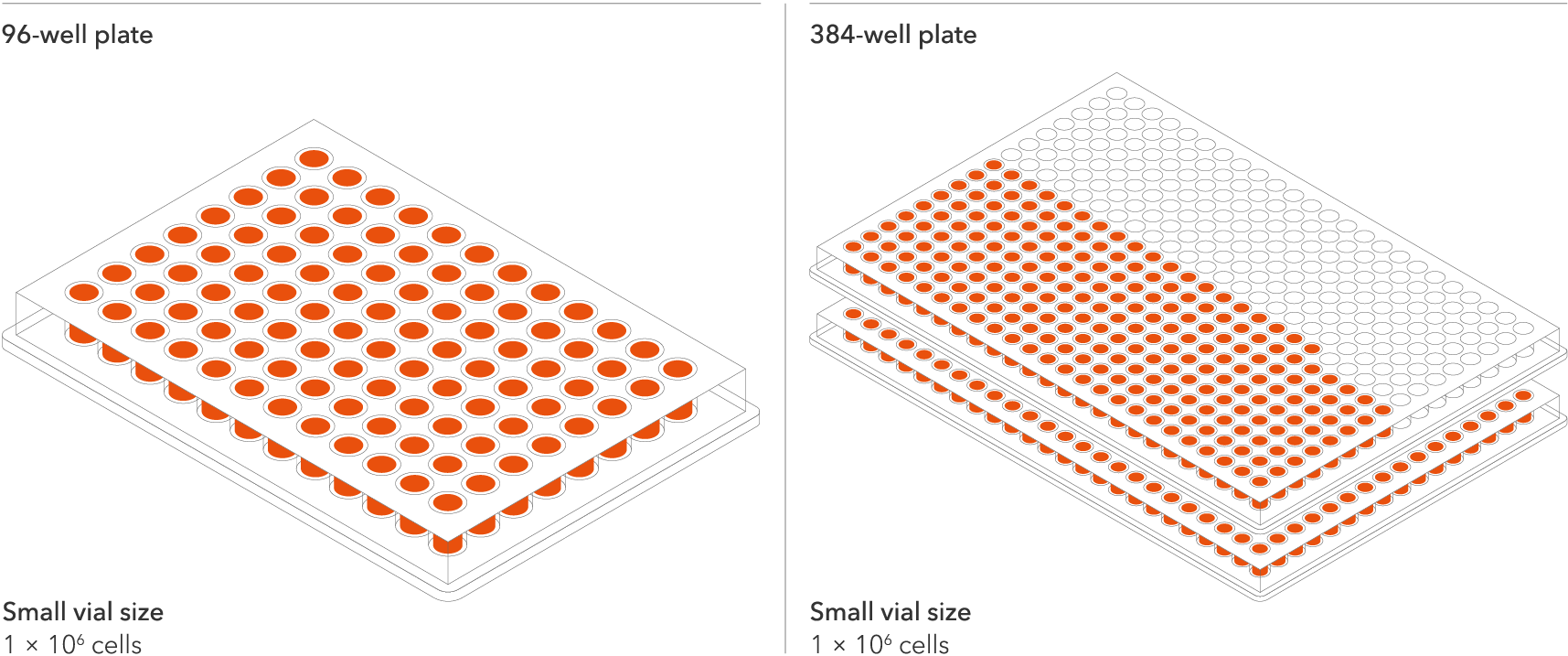
Industry leading seeding density
The recommended minimum seeding density is 30,000 cells/cm2, compared to up to 250,000 cells/cm2 for other similar commercially available products. One small vial can plate a minimum of 0.7 x 24-well plate, 1 x 96-well plate, or 1.5 x 384-well plates. This means every vial goes further, enabling more experimental conditions and more repeats, resulting in more confidence in the data.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.





Hoescht(blue)_day12v2.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_20xMAP2(red)Hoescht(blue)_day12v2.png)
Hoescht(blue)TUBB3(blue)_day4.jpg?width=604&name=bit.bio_ioGlutamatergic%20Neurons_60xMAP2(red)Hoescht(blue)TUBB3(blue)_day4.jpg)

.png?width=1860&height=1260&name=bit.bio_3x2_ioGlutamatergic%20Neurons_MAP2_Hoescht_x20_hi.res%20(1).png)











