



























cat no | io1027
ioMotor Neurons
Human iPSC-derived motor neurons
-
Cryopreserved human iPSC-derived cells powered by opti-ox, that are ready for experiments in days
-
Ideal for modelling ALS, neurodegeneration research, and neuromuscular disease studies
-
Clump free, highly-pure motor neurons, that form functional neuronal networks in co-culture with astrocytes

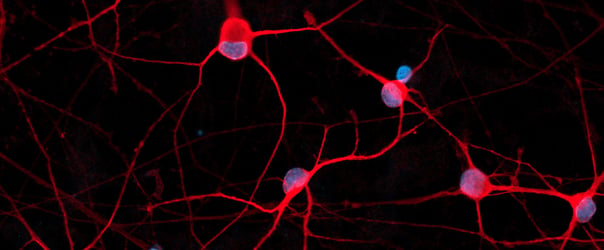
Human iPSC-derived motor neurons

ioMotor Neurons acquire a rapid motor neuronal phenotype, without clumping
Time-lapse video capturing the rapid and homogeneous motor neuronal phenotype acquisition upon thawing of cryopreserved ioMotor Neurons, showing no signs of clumping. 11 day time course.

Functional neuronal networks are detected in astrocyte co-culture from day 14
ioMotor Neurons are functional – showing activity in astrocyte co-culture that increases over time as networks mature. Mean firing rates (electrode spike count divided by the total time of the recording period) is shown to increase substantially throughout the course of the experiment, as demonstrated by multielectrode array activity (MEA).

Functional neuronal networks are detected in astrocyte co-culture from day 14
Spontaneous neuronal activity is exhibited from as early as day 14 and continues to increase up to the final measured timepoint, day 42.

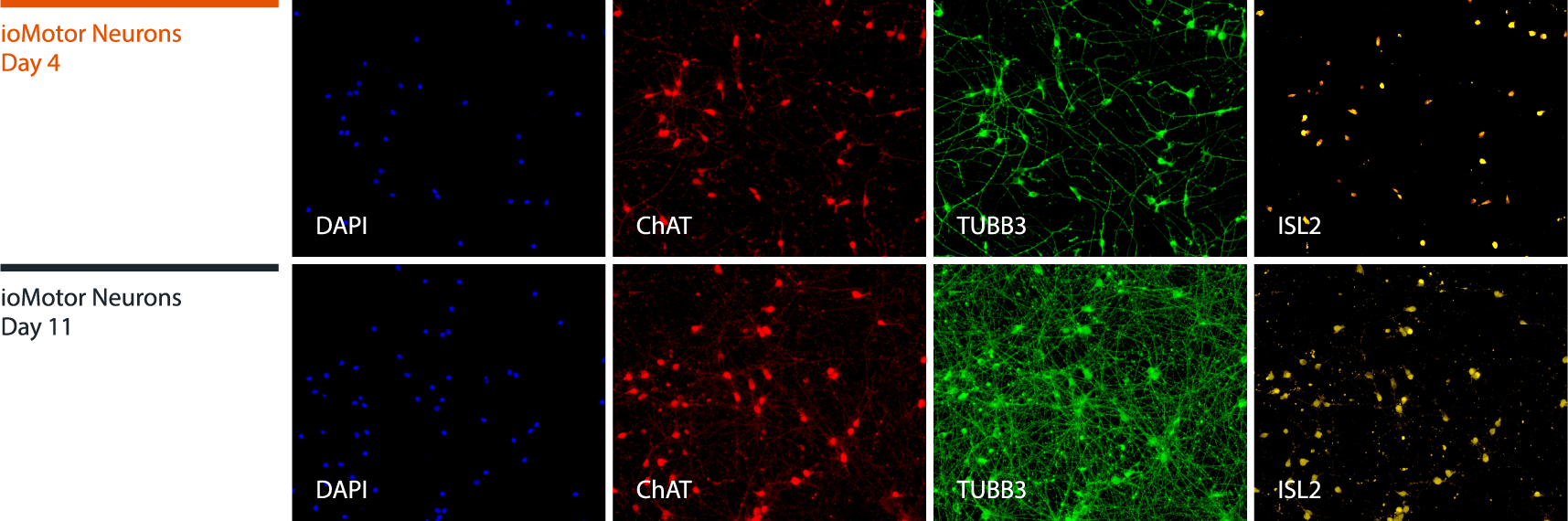
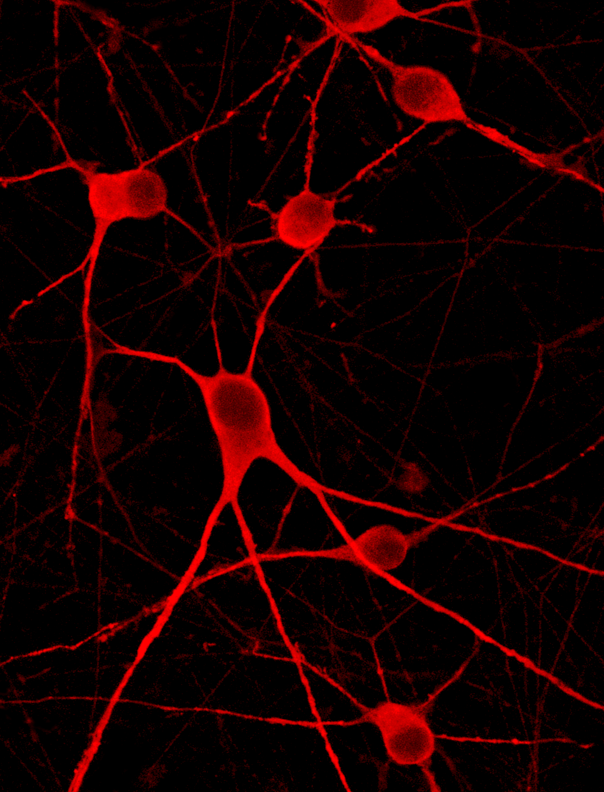
Immunocytochemistry shows protein expression of key motor neuron markers
Immunofluorescent staining on post-revival day 4 and day 11 demonstrates homogenous expression of the pan-neuronal protein TUBB3, motor neuron specific marker ISL2, the cholinergic marker ChAT and nuclear staining (DAPI).

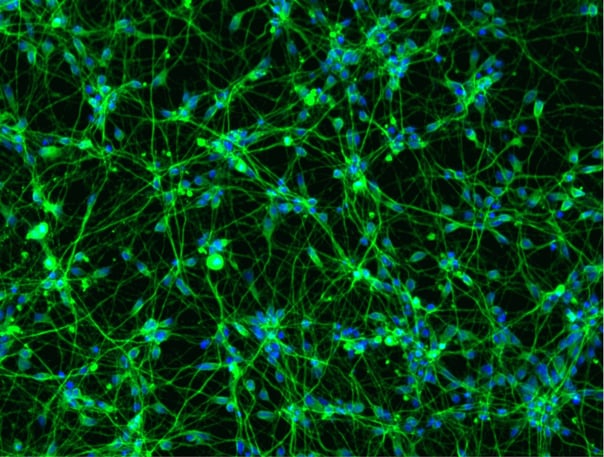
Immunocytochemistry shows protein expression of key motor neuron markers
Immunofluorescent staining on post-revival day 4 and day 11 demonstrates homogenous expression of the pan-neuronal protein MAP2, motor neuron specific marker HB9, the cholinergic marker VAChT and nuclear staining (DAPI).

RT-qPCR shows gene expression of key motor neuron markers
RT-qPCR gene expression on post-revival days 1, 4, 11 & 18 demonstrates rapid acquisition of motor neuron genotype, shown by the expression of pan-neuronal, cholinergic & key lower motor neuron markers from as early as day 1. Pluripotency markers NANOG and OCT4 are swiftly downregulated.

Bulk RNA-sequencing exhibits a HOX gene signature indicative of a spinal motor neuron (cervical region) identity
Expression of HOX genes was evaluated using bulk RNA sequencing data. This heatmap shows expression of genes from the B cluster and expression of HOXC4 and HOXC5, although at lower levels. This data, together with the marker expression from single cell RNA sequencing, suggests that ioMotor Neurons have a spinal cord (cervical region) identity. Note, this data is from cells in continuous culture and not cryopreserved cells.

Single cell RNA-sequencing shows ioMotor Neurons form a pure population (>99.9%) of neurons
Single cell RNA-sequencing analysis was performed with ioMotor Neurons at four timepoints: day 0 (iPSCs), 4, 7, and 14. Gene expression was assessed by 10x Genomics single cell RNA-sequencing. Note, this data is from cells in continuous culture and not cryopreserved cells. By day 14, the population has a distinct expression profile indicating a pure population (>99.9%) of post-mitotic neurons, demonstrated through the expression pan-neuronal markers MAP2 and TUBB3.

Single cell RNA-sequencing shows ioMotor Neurons express key spinal motor neuron markers, >80% of cells express MNX1 on day 14
Starting from day 4, the expression of the key spinal motor neuron marker genes MNX1 (HB9), FOXP1, and ISL2 is detected in the culture, with >80% of cells expressing MNX1 on day 14. These percentages are likely to be an underestimation due to limitation of single cell RNA sequencing, as ICC for HB9 & ISL2 shows homogeneous expression of these markers in our cultures.

Single cell RNA-sequencing shows a high proportion of ioMotor Neurons express cholinergic markers by day 7
Within 7 days, the expression of the key cholinergic marker genes CHAT & SLC18A3 (VAChT) are detected in a high proportion of ioMotor Neurons.

Bulk RNA-sequencing demonstrates high batch-to-batch consistency of ioMotor Neurons
Bulk RNA sequencing analysis was performed on three independent batches of ioMotor Neurons at three different time points throughout the reprogramming protocol. Principal component analysis represents the variance in gene expression between the batches of ioMotor Neurons. This analysis shows high consistency between each batch of ioMotor Neurons at each given timepoint. Populations of ioMotor Neurons with equivalent expression profiles can be generated consistently from every vial, allowing confidence in experimental reproducibility. Note, this data is from cells in continuous culture and not cryopreserved cells.

Co-culture of ioMotor Neurons and ioSkeletal Myocytes
High resolution confocal imaging of ioSkeletal Myocytes (io1002) and ioMotor Neurons co-culture. Staining with alpha-bungarotoxin (yellow) highlights acetylcholine receptor expression on co-cultured ioSkeletal Myocytes. Desmin (cyan) and microtubule-associated protein 2 (red) define ioSkeletal Myocytes and ioMotor Neurons respectively. Co-culture imaged at day 30, 40X magnification.
Download the step-by-step protocol for culturing ioSkeletal Myocytes and ioMotor Neurons.

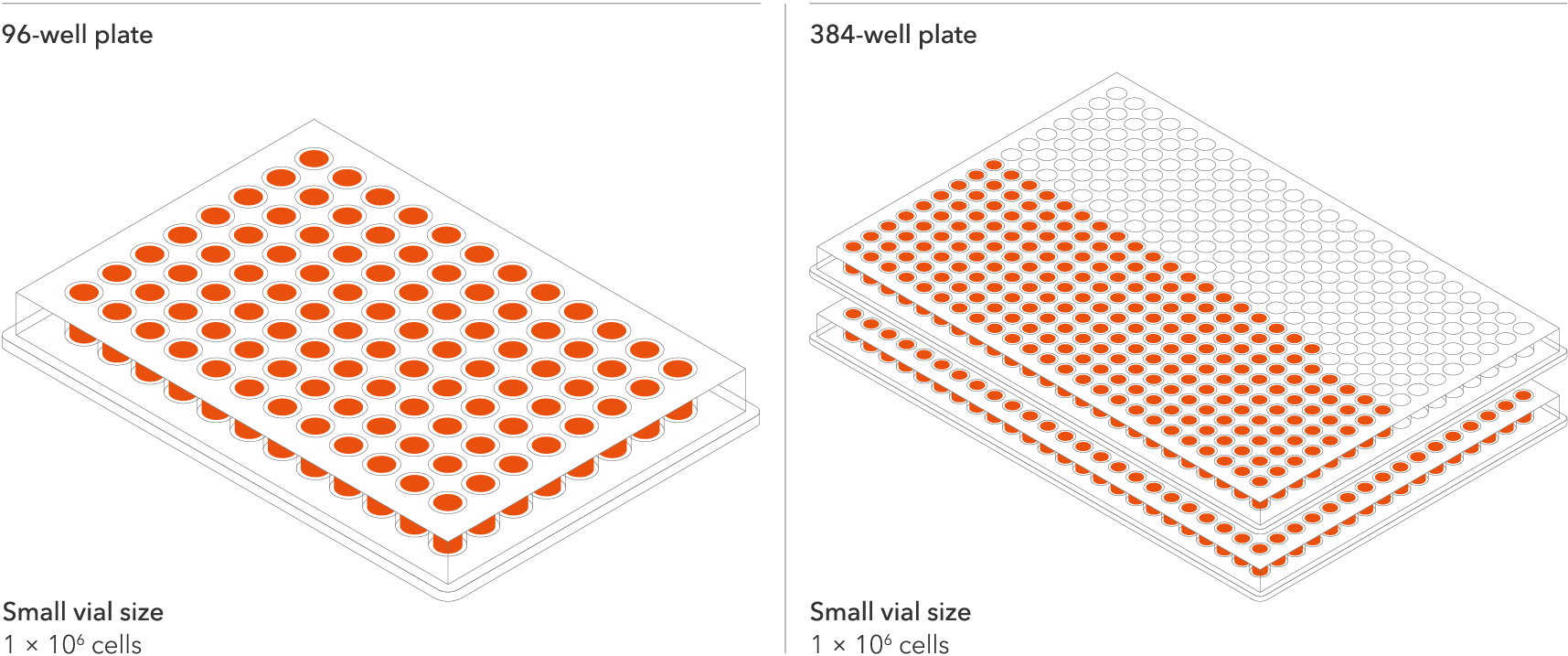
Do more with every vial
The seeding density of our human iPSC-derived ioMotor Neurons and related disease models has been optimised and validated to a recommended seeding density of 30,000 cells/cm2. This means scientists can do more with every vial and expand experimental design within budget without losing out on quality. Resulting in more experimental conditions, more repeats, and more confidence in the data. One Small vial can plate a minimum of 0.7 x 24-well plate, 1 x 96-well plate, or 1.5 x 384-well plate.

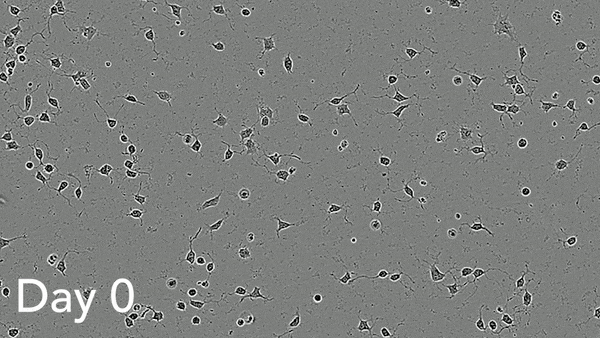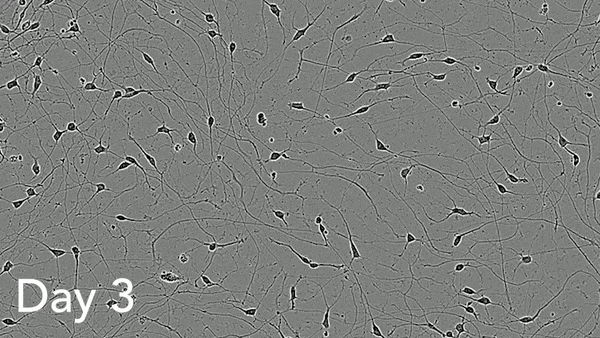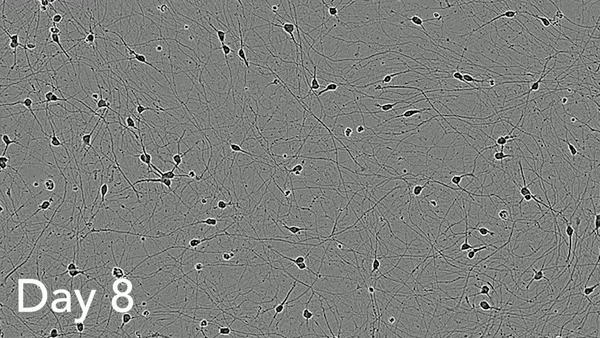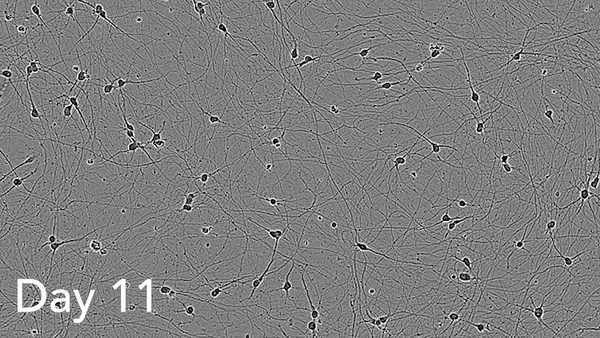
ioMotor Neurons form a homogenous neuronal network by day 4
ioMotor Neurons mature rapidly and form homogenous populations over 18 days. Day 1 to 18 post thawing; 100X magnification.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.






Hoescht(blue)TUBB3(blue)_day4.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_60xMAP2(red)Hoescht(blue)TUBB3(blue)_day4.png)

.png?width=604&name=Tech%20Nets%20online%20symposium%20header%20(1).png)






.png?width=604&name=ioMotor%20Neuron%20Hero%20image%20for%20Webinar%20(1).png)












/ioSkeletal-Myocytes-DMD-Exon-52-Deletion-hero.webp?width=1276&height=1190&name=ioSkeletal-Myocytes-DMD-Exon-52-Deletion-hero.webp)


