



















cat no | io1024
ioSensory Neurons
Highly pure human iPSC-derived sensory neurons with a defined nociceptor identity
-
Cryopreserved human iPSC-derived cells powered by opti-ox, that are ready for experiments in days
-
Ideal for chronic pain research, drug development, and neurotoxicology
-
Display spontaneous neuronal activity and exhibit a functional nociceptor phenotype

Highly pure human iPSC-derived sensory neurons with a defined nociceptor identity
opti-ox programmed ioSensory Neurons rapidly form a homogenous neuronal population
Time-lapse video capturing the rapid and homogeneous neuronal phenotype acquisition upon thawing of cryopreserved ioSensory Neurons. 14 day time course.

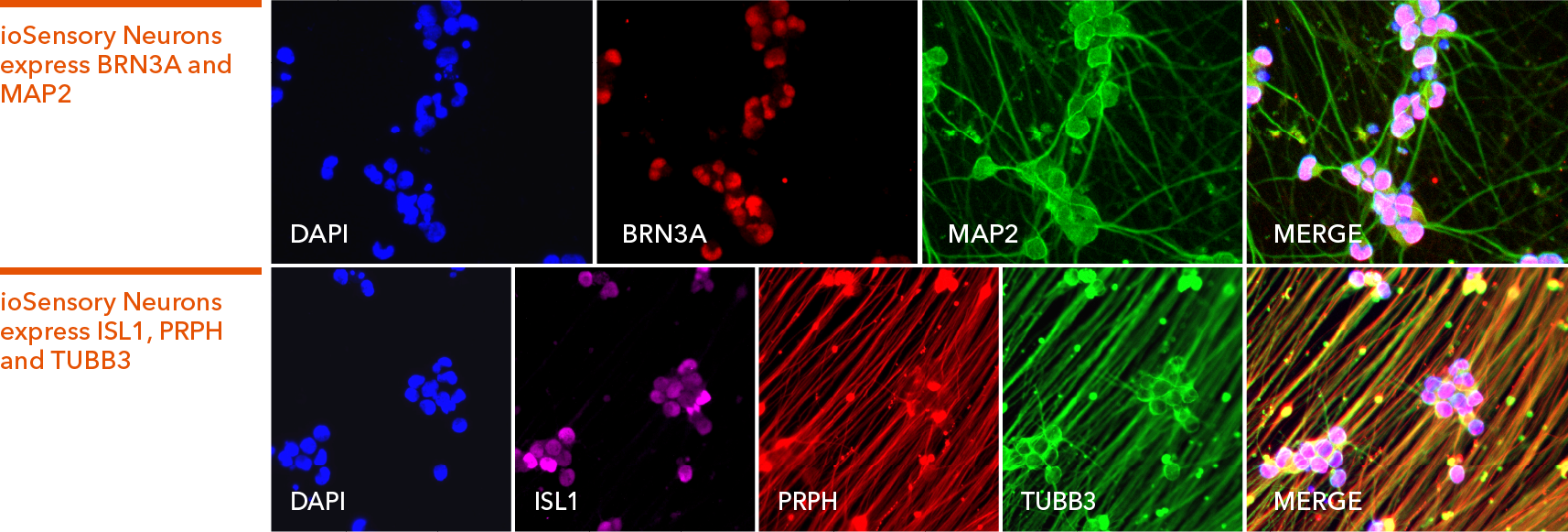
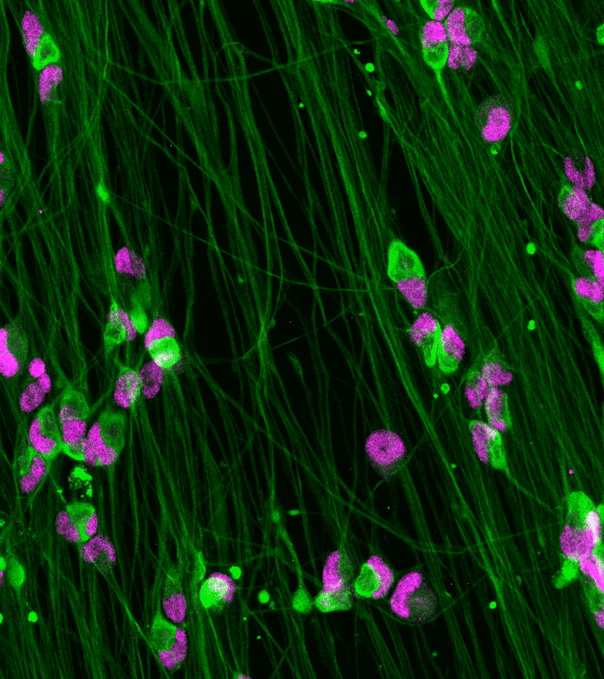
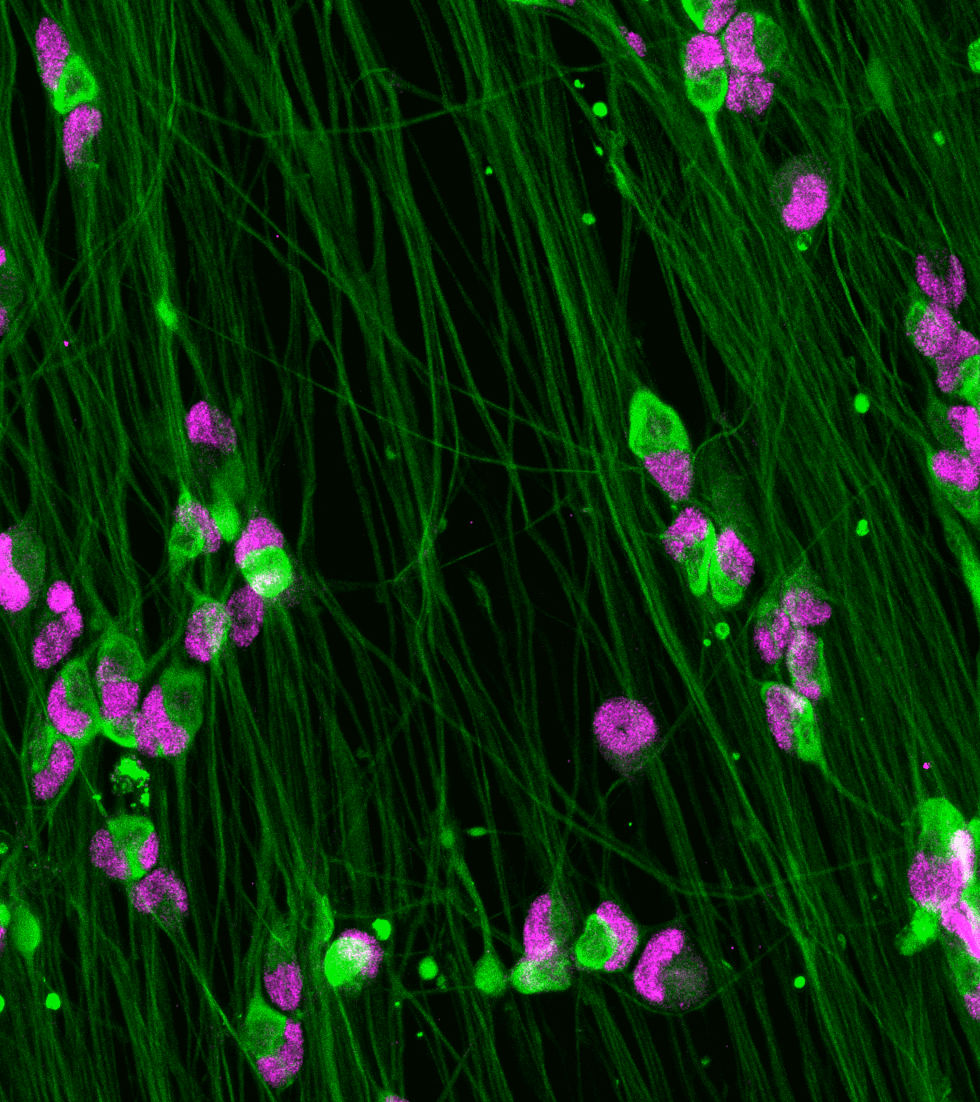
ioSensory Neurons express pan-sensory neuron-specific markers
Immunofluorescent staining of ioSensory Neurons at day 14 post-revival. The upper panel shows that ioSensory Neurons are positive for BRN3A (red), the pan-neuronal marker MAP2 (green), and the DAPI counterstain (blue), and demonstrates that all MAP2 positive neurons have a sensory neuronal identity. The lower panel shows that ioSensory Neurons are positive for ISL1 (magenta), PRPH (red), the pan-neuronal marker TUBB3 (green), and the DAPI counterstain (blue).

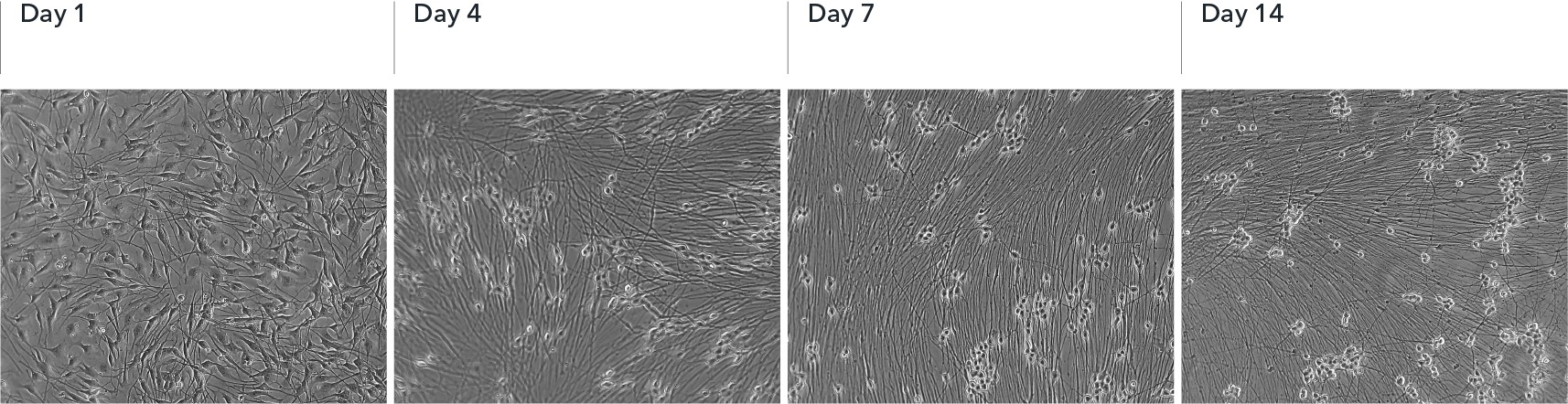
ioSensory Neurons show visible neuronal networks by day 7
Upon deterministic programming, rapid morphological changes are observed in the cells, with neurons identified by day 4 post-revival. Visible neuronal networks are observed by day 7 post-thaw. Images show day 1 to 14 post-thawing; 10X magnification.

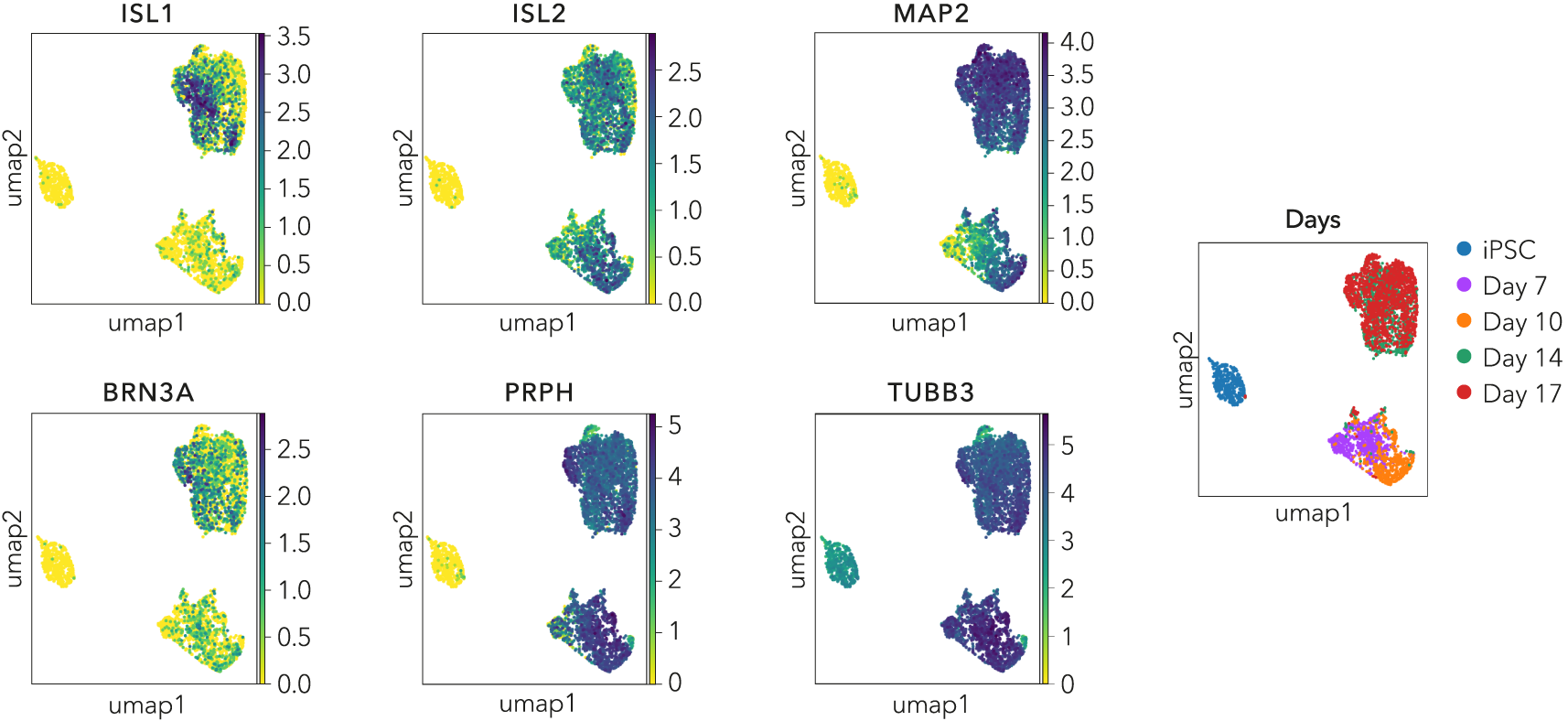
Single cell RNA-sequencing shows ioSensory Neurons form a pure population (>99%) of sensory neurons and express key pan-sensory neuronal markers
Single cell RNA-sequencing analysis was performed with ioSensory Neurons at five specific timepoints: day 0 (iPSCs), 7, 10, 14 and 17. By day 7, the population has a distinct expression profile indicating a pure population (>99%) of post-mitotic sensory neurons, demonstrated through the expression of pan-sensory neuronal marker genes, ISL1, ISL2, PRPH, and BRN3A, together with the pan-neuronal markers TUBB3 and MAP2. Gene expression was assessed by 10x Genomics single cell RNA-sequencing. Note, this data is from cells in continuous culture, so minor variations may exist between this data and data from cryopreserved cells.

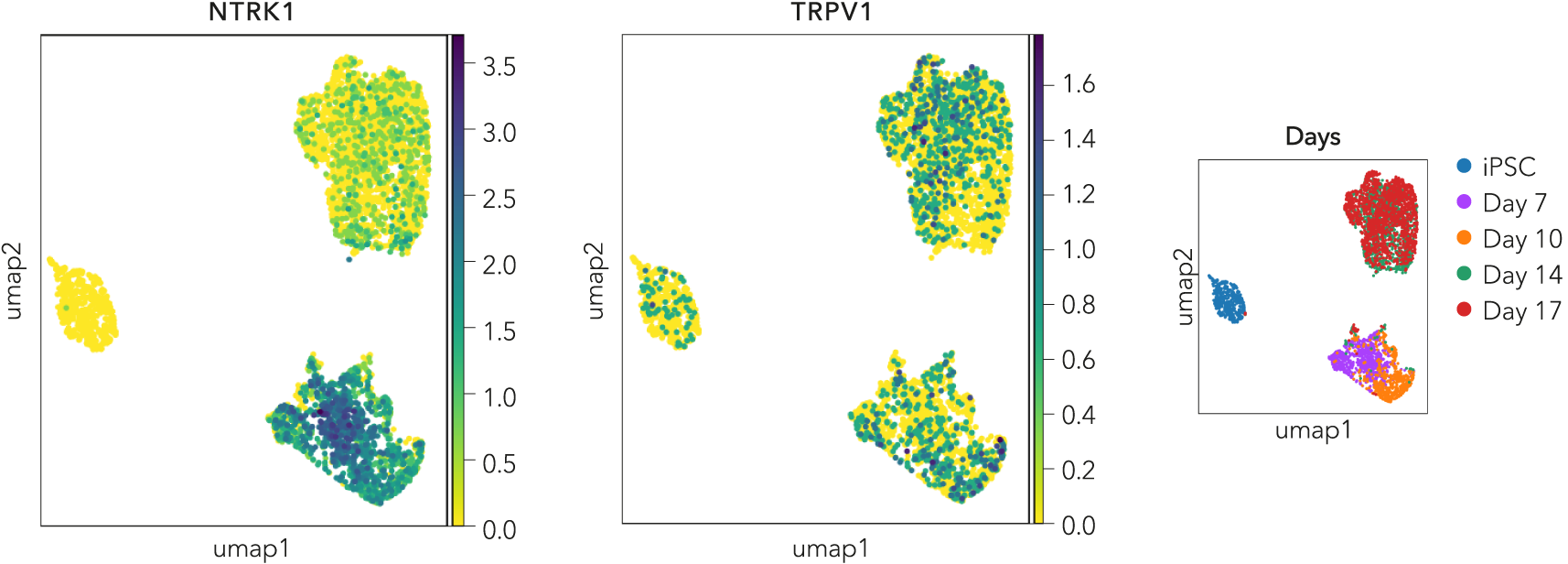
Single cell RNA-sequencing shows ioSensory Neurons express key nociceptor markers
Single cell RNA-sequencing analysis was performed with ioSensory Neurons at five specific timepoints: day 0 (iPSCs), 7, 10, 14 and 17. By day 7, the expression of key nociceptor marker genes (NTRK1 and TRPV1) could be detected in post-mitotic sensory neurons. Gene expression was assessed by 10x Genomics single cell RNA-sequencing. Note, this data is from cells in continuous culture, so minor variations may exist between this data and data from cryopreserved cells.

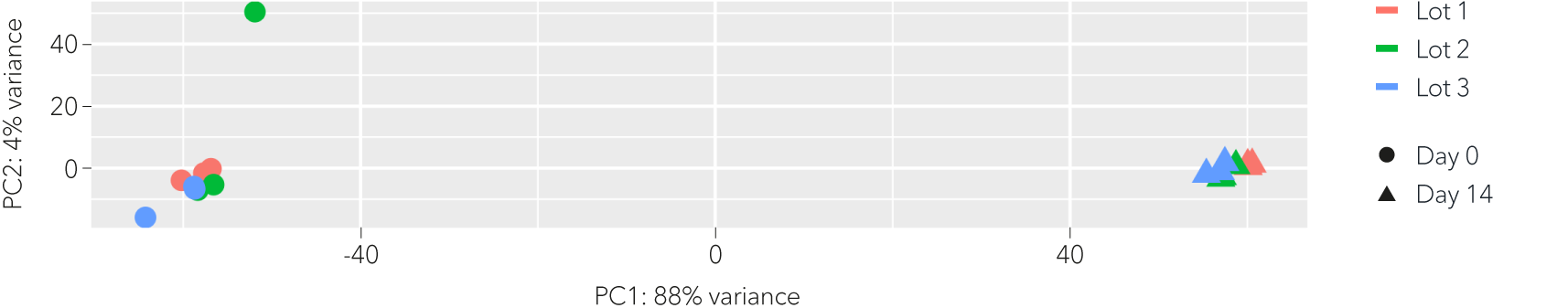
Whole transcriptome analysis shows high lot-to-lot consistency of ioSensory Neurons
Bulk RNA sequencing analysis was performed on three technical replicates of three independent lots of ioSensory Neurons at different time points throughout the reprogramming protocol. Principal component analysis represents the variance in gene expression between the lots of ioSensory Neurons. This analysis shows high consistency between each lot of ioSensory Neurons at each given timepoint. Differential gene expression was found to be less than 1% between lots, at day 14 post-thaw. Pure populations of ioSensory Neurons with equivalent expression profiles can be generated consistently from every vial, allowing confidence in experimental reproducibility. Expression levels for specific genes of interest can be requested by contacting our team at technical@bit.bio.

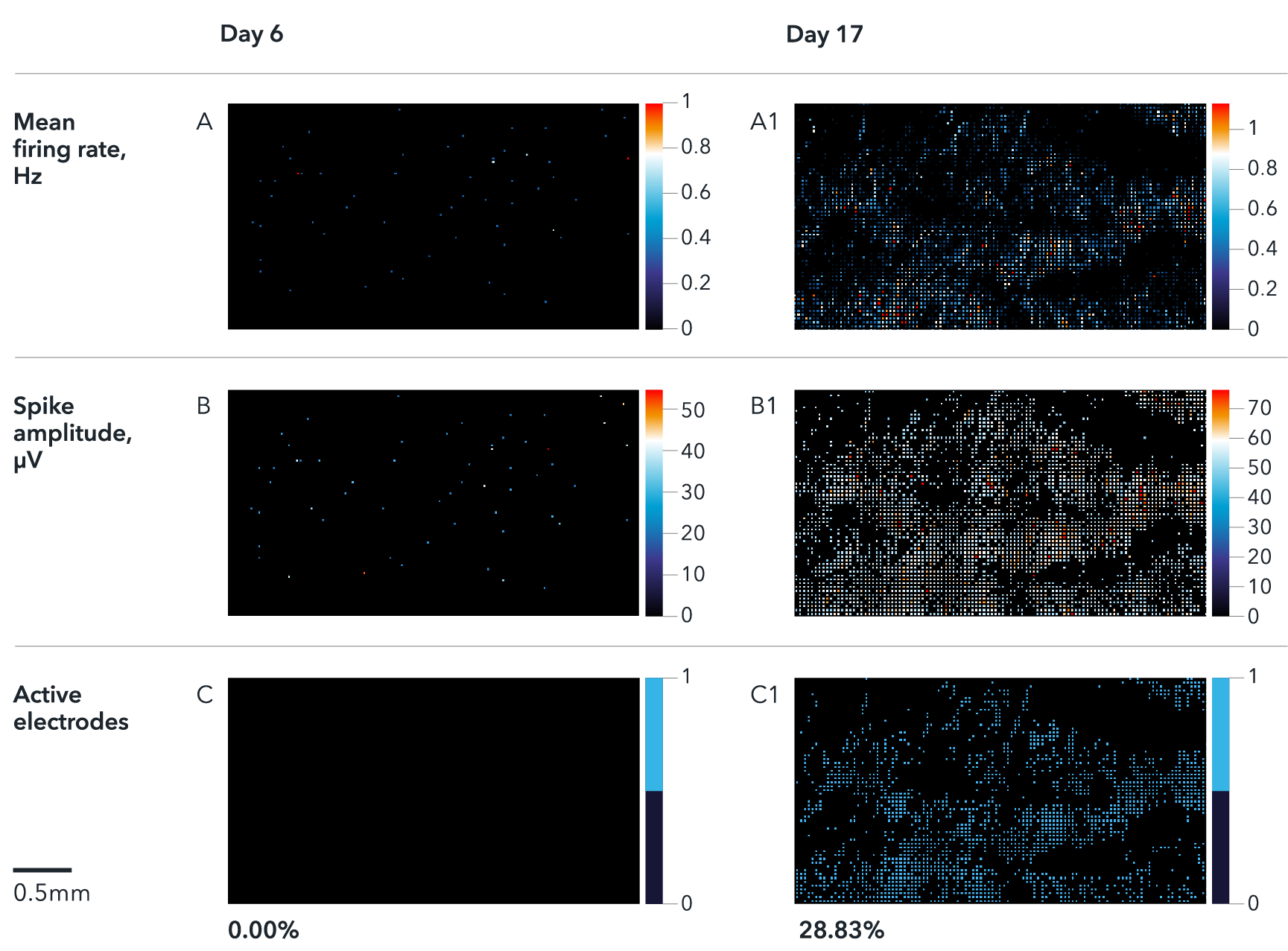
ioSensory Neurons display spontaneous activity that matures over time
Multi electrode array (MEA) recordings of ioSensory Neurons at days 6 and 17. The activity maps show firing rate (A), spike amplitude (B) and % of active electrodes (C). Results demonstrate a time-dependent increase of spontaneous activity during neuronal maturation from day 6 to day 17. Analysis was performed on a Maxwell Biosystem's MaxTwo multi-well system. Note, this data is from cells in continuous culture and not cryopreserved cells.

ioSensory Neurons display a functional nociceptor phenotype
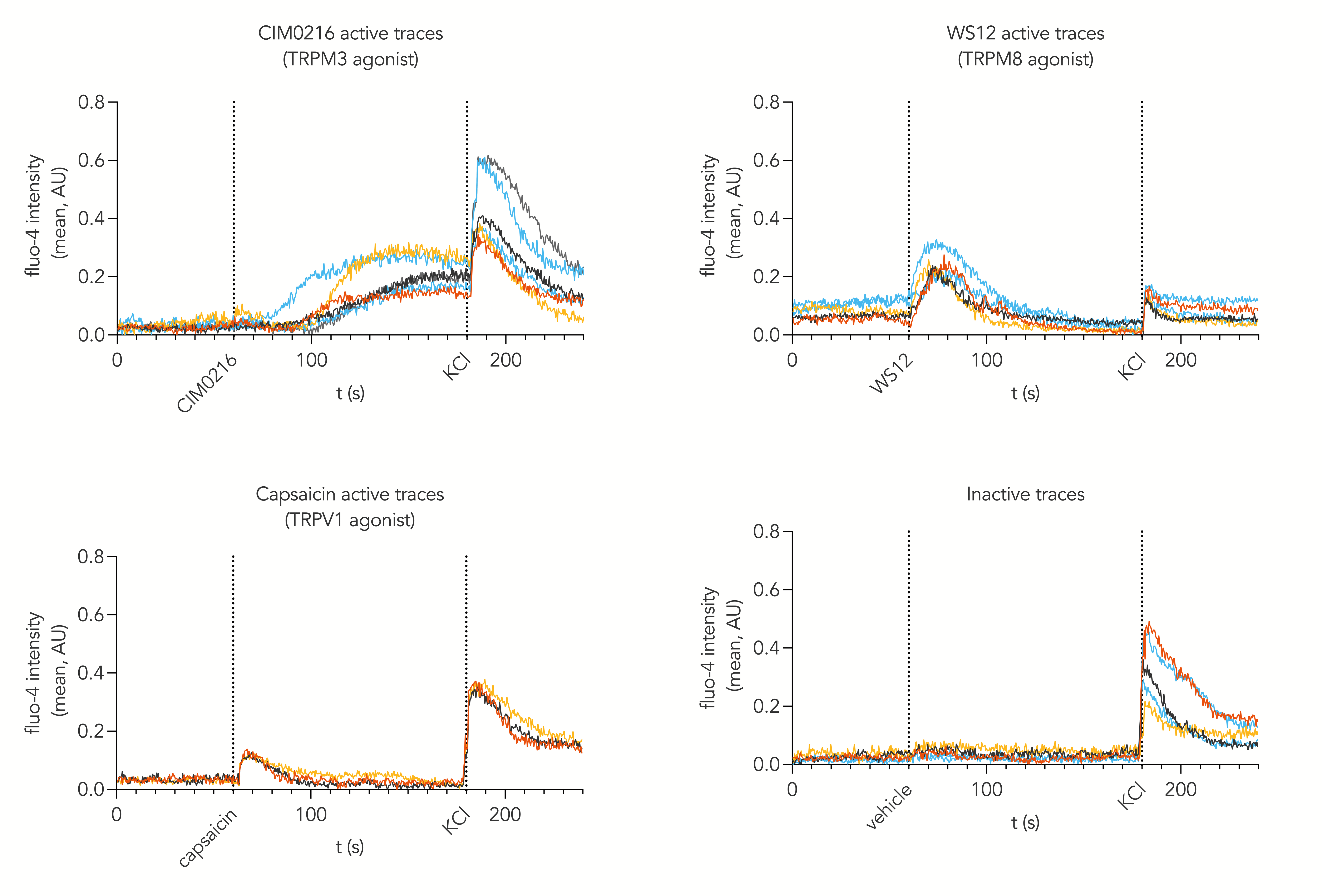
Calcium mobilisation imaging upon stimulation of ioSensory Neurons with pharmacological agonists targeting thermosensitive TRP channels such as TRPV1 (capsaicin), TRPM3 (CIM0216) and TRPM8 (WS12). Active traces represent the increase in intracellular calcium mobilisation of individual cells upon exposure to noxious stimuli but not to vehicle at day 14 post-revival. This indicates that cells display a functional nociceptor phenotype within 14 days post-thaw.
Calcium imaging of ioSensory Neurons demonstrates activity following treatment with a TRPM3 agonist
Representative video of ioSensory Neurons displaying calcium mobilisation in response to treatment with the TRPM3 agonist, CIM0216. Note, this data is from cells in continuous culture and not cryopreserved cells.

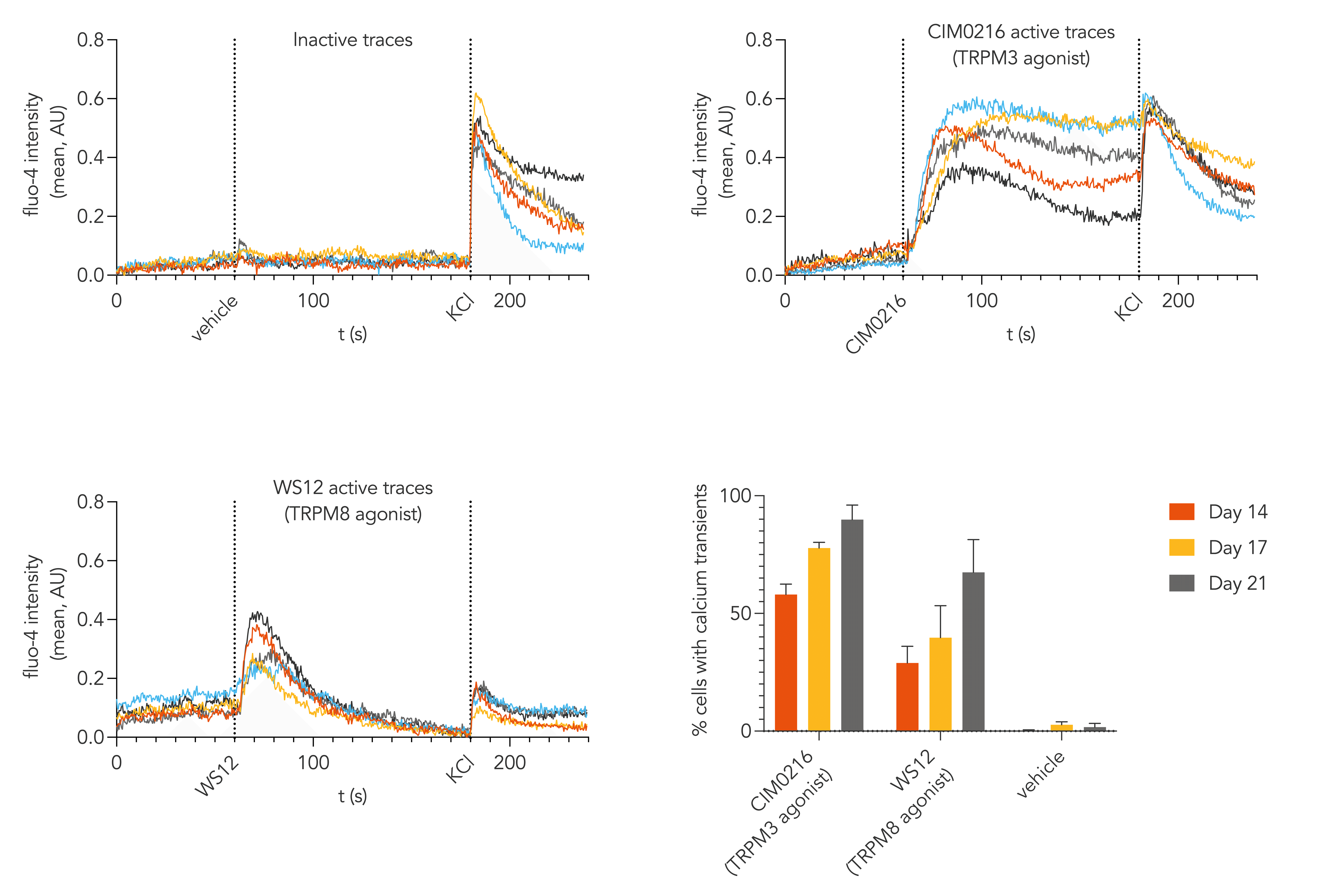
ioSensory Neurons display enhanced TRPM3 and TRPM8 responses with bespoke media for these channels and increased culture length
Calcium mobilisation imaging, performed with ioSensory Neurons cultured using a bespoke media up to day 14, 17 or 21 post-thaw, shows that ioSensory Neurons display strong responses to pharmacological agonists targeting thermosensitive TRP channels, TRPM3 (CIM0216) and TRPM8 (WS12). The top and bottom left graphs show active traces which represent the increase in intracellular calcium mobilisation of individual cells, at day 21 post-thaw, upon exposure to noxious agonists but not to vehicle, indicating that cells display features of functional nociceptors. The bottom right graph shows the percentage of responding cells at day 14, 17, or 21 post-thaw – the bespoke media and increased culture length appears to greatly enhance the percentage of cells responding to the TRPM3 and TRPM8 agonists, to 90% and 67% at day 21 post-thaw, respectively. Please contact us at technical@bit.bio for further information on the bespoke media.

Automated patch clamp compatibility and clear functional expression of TTX resistant sodium channels
Medium-throughput automated patch clamp analysis (Patchliner, Nanion Technologies) recordings of Day 21 ioSensory Neurons showing evidence of sodium currents in the presence of TTX. TTX resistant sodium channels include NaV1.8, NaV1.9 or NaV1.5. The highest concentration of 100 nM blocked the majority of sodium currents, but some functional TTX-resistant sodium current remained The IC50 of TTX was approximately 7.5 nM (n = 5).
Data collected and analysed by Nanion Technologies. More information can be found in this application note written by Nanion.

Heat responsiveness during patch clamp recordings
ioSensory Neurons demonstrate a clear heat response in medium-throughput automated patch clamp analysis (Patchliner, Nanion Technologies). Day 21 ioSensory Neurons showed a 2-fold increase in inward (blue) currents in response to culture media heated to 45°C, indicating functional expression of heat responsive channels including TRPV1 and TRPM3.
Data collected and analysed by Nanion Technologies. More information can be found in this application note written by Nanion.

ioSensory Neurons are efficiently transfected with mRNA encoding GFP
ioSensory Neurons are efficiently transfected and show sustained long-term expression of mRNA encoding GFP. ioSensory Neurons were imaged from Day 1 post-thaw and throughout the experiment to assess transfection efficiency and evaluate potential cytotoxic effects of the transfection protocol. Day 1 images were captured prior to transfections on the same day.
Download the step-by-step protocol for lipid-based delivery of synthetic mRNA into ioSensory Neurons.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.

.jpg?width=713&height=549&name=Ryan%20jones%20headshot%20(2).jpg)