


.png?width=1715&height=651&name=ioGlutamatergic_neurons-CRISPR-Morphology%20(1).png)
%20compressed.png?width=1715&height=1221&name=ioGlutamatergic_neurons-CRISPR-ICC%20(1)%20compressed.png)





.png?width=1715&height=651&name=ioGlutamatergic_neurons-CRISPR-Morphology%20(1).png)
%20compressed.png?width=1715&height=1221&name=ioGlutamatergic_neurons-CRISPR-ICC%20(1)%20compressed.png)


cat no | io1090
CRISPRko-Ready ioGlutamatergic Neurons
Human iPSC-derived glutamatergic neurons expressing Cas9 for rapid gene knockout generation
- Cryopreserved human iPSC-derived cells powered by opti-ox that are ready for experiments in days
- Neurons for functional genomics, stably expressing Cas9 for gene knockouts and CRISPR screens
- Consistent, functional excitatory neurons that form neuronal networks within days

Human iPSC-derived glutamatergic neurons expressing Cas9 for rapid gene knockout generation
Amplicon sequencing demonstrates high knockout efficiency of SOX11 by both lentiviral transduction and lipid-based transfection
SOX11 indel formation was measured by amplicon sequencing in CRISPRko-Ready ioGlutamatergic Neurons, that were either transfected or transduced with a gRNA targeting SOX11. gRNAs were introduced into the cells 1 or 3 days after thawing using lentiviral transduction or synthetic gRNA delivery with Lipofectamine RNAiMAX transfection reagent. After 3 days of culture following guide delivery, DNA was harvested for amplicon sequencing of SOX11. Comparable knockout efficiencies were achieved with both methods of gRNA delivery. A non-targeting gRNA was used as a control.
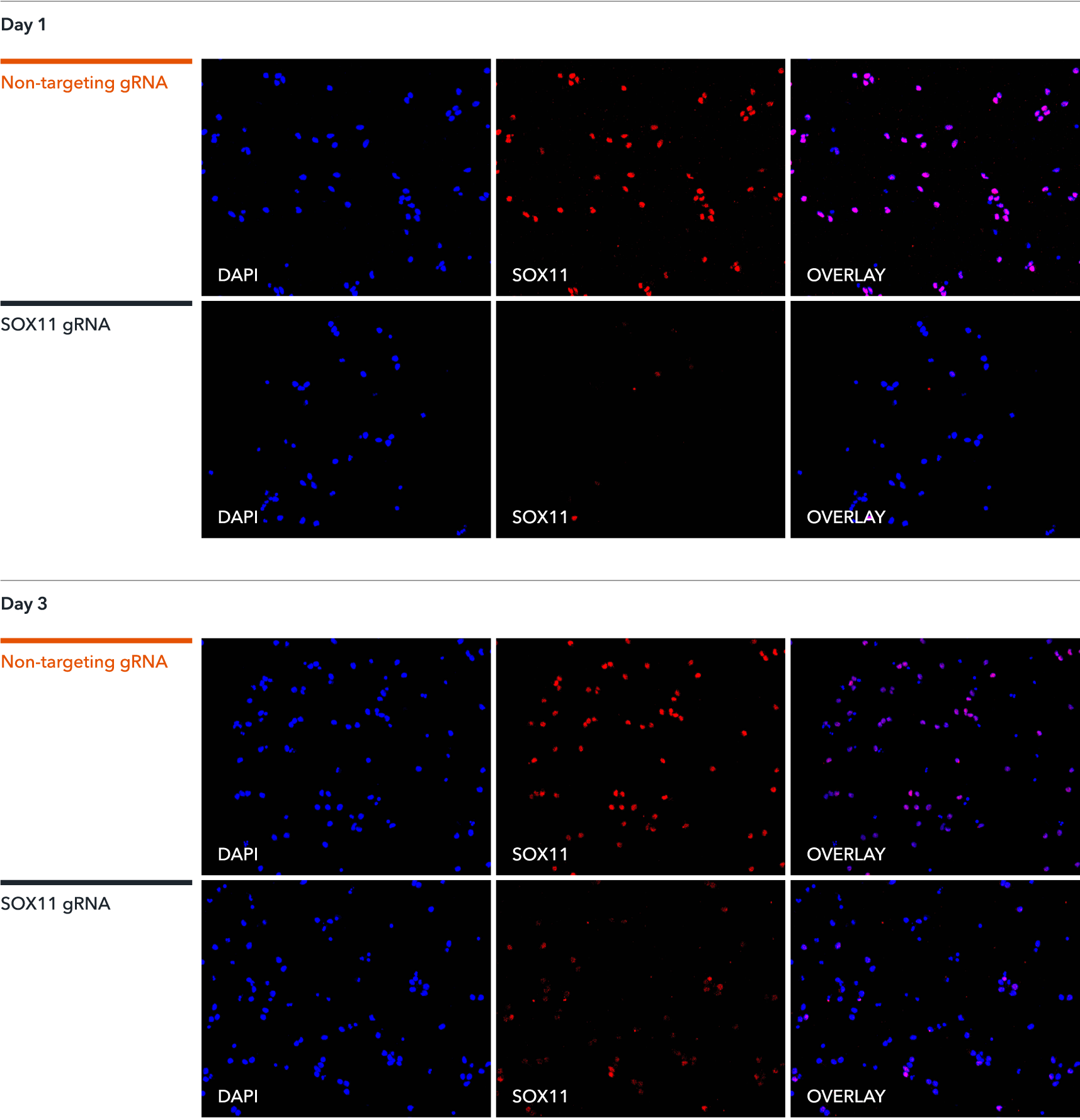
Immunofluorescence staining demonstrates high knockout efficiency of SOX11 by lentiviral transduction
Immunofluorescence staining of CRISPRko-Ready ioGlutamatergic Neurons demonstrates a highly efficient knockout of SOX11. The gRNAs were delivered by lentiviral transduction or transfection of synthetic gRNA using Lipofectamine RNAiMAX on day 1 or day 3 post-revival. Immunofluorescence staining of SOX11 was conducted five days post gRNA delivery. Similar knockout efficiencies were achieved for both methods of gRNA delivery. A non-targeting gRNA was used as a control.

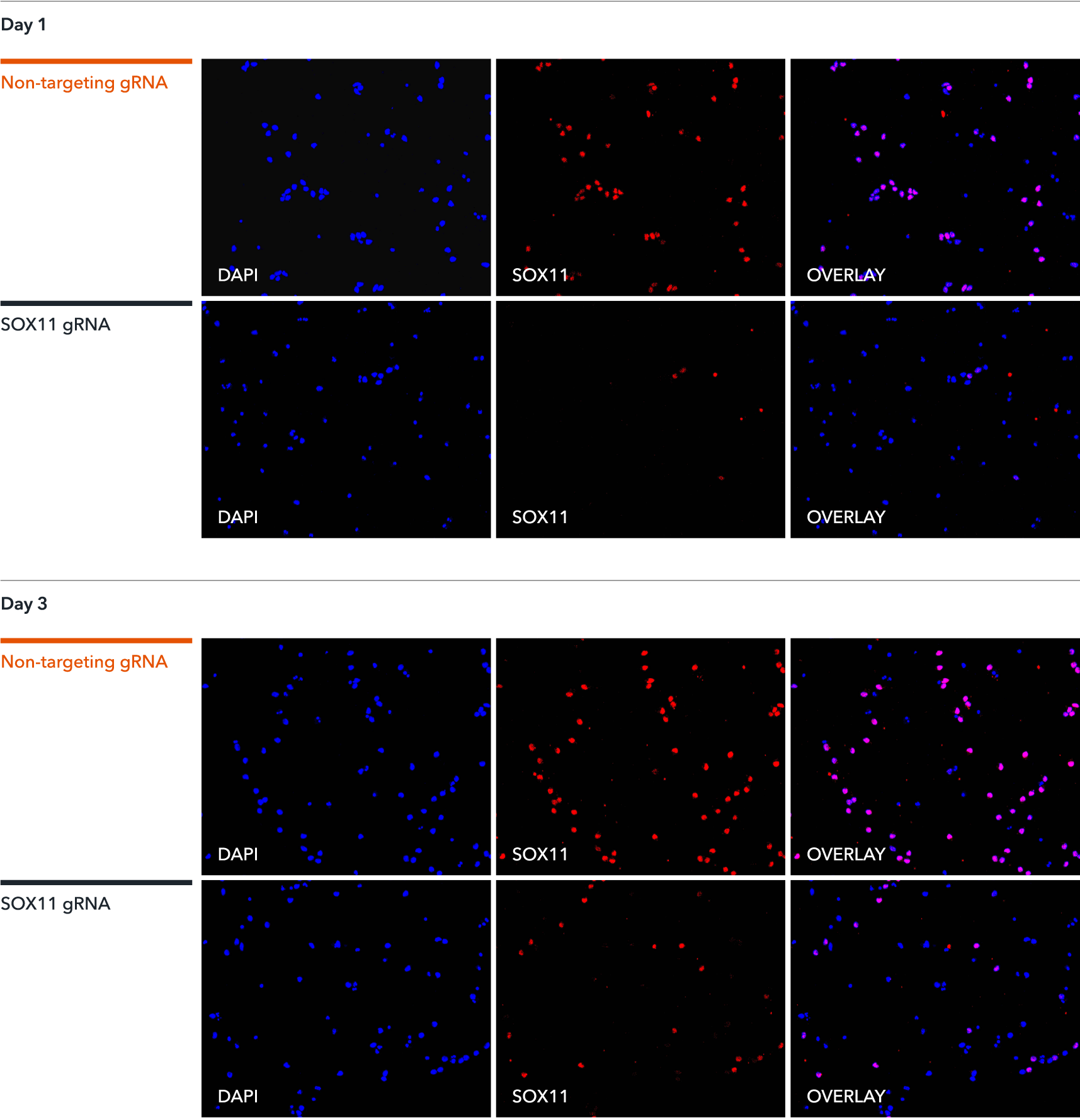
Immunofluorescence staining demonstrates high knockout efficiency of SOX11 by lipid-based transfection
Immunofluorescence staining of CRISPRko-Ready ioGlutamatergic Neurons demonstrates a highly efficient knockout of SOX11. The gRNAs were delivered by lentiviral transduction or transfection of synthetic gRNA using Lipofectamine RNAiMAX on day 1 or day 3 post-revival. Immunofluorescence staining of SOX11 was conducted five days post gRNA delivery. Similar knockout efficiencies were achieved for both methods of gRNA delivery. A non-targeting gRNA was used as a control.

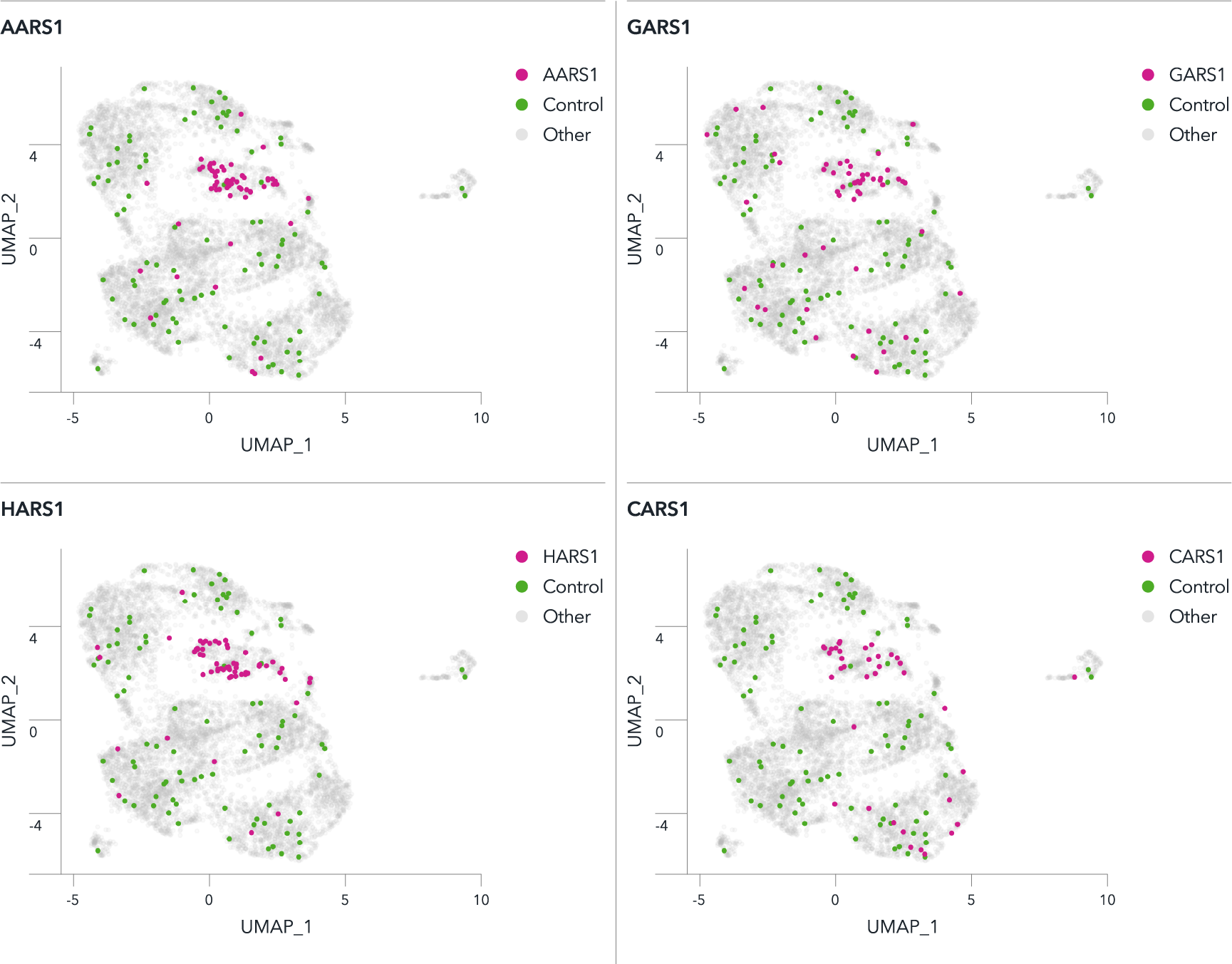
A pooled knockout screen of neurodegenerative disease-relevant genes in CRISPRko-Ready ioGlutamatergic Neurons shows clustering of aaRS genes in UMAPs
For a pooled knockout screen in CRISPRko-Ready ioGlutamatergic Neurons, 100 known genes involved in neurodegenerative diseases were selected. Lentiviral transduction of the gRNAs was carried out on day 3 and single-cell gene expression analysis was performed on day 12. Single cells were clustered on uniform manifold approximations and projections (UMAPs) based on their shared nearest neighbour’s gene expression. Clustering of aminoacyl-tRNA synthetase (aaRS) knockouts including AARS1, HARS1, CARS1, and GARS1 was observed. In contrast, cells transduced with non-targeting control sgRNAs were evenly distributed among clusters. Pathway analysis showed gRNAs targeting aaRSs activated the unfolded protein response (UPR), the mechanism by which cells control endoplasmic reticulum protein homeostasis. In many neurodegenerative diseases, signs of UPR activation have been reported. The most common aaRS-associated monogenic disorder is the incurable neurodegenerative disease Charcot–Marie–Tooth neuropathy (CMT).
.png?width=1715&height=651&name=ioGlutamatergic_neurons-CRISPR-Morphology%20(1).png)
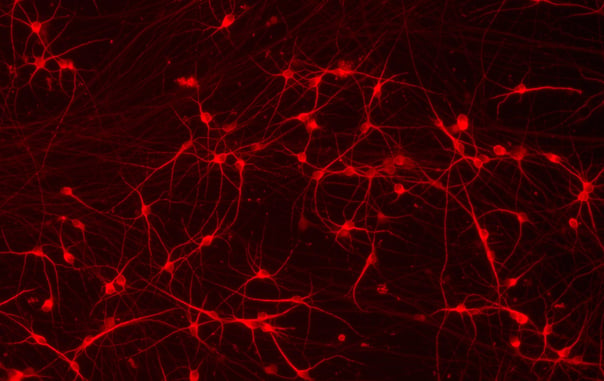
CRISPRko-Ready ioGlutamatergic Neurons form structural neuronal networks by day 11
CRISPRko-Ready ioGlutamatergic Neurons mature rapidly, show glutamatergic neuron morphology and form structural neuronal networks over 11 days, highly similar to wild-type ioGlutamatergic Neurons (io1001). Day 1 to 11 post-thawing; 100X magnification.
%20compressed.png?width=1715&height=1221&name=ioGlutamatergic_neurons-CRISPR-ICC%20(1)%20compressed.png)
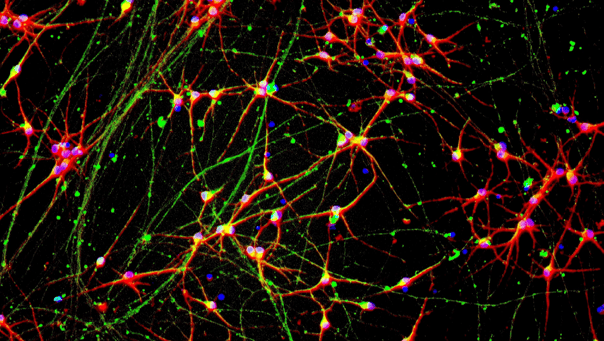
CRISPRko-Ready ioGlutamatergic Neurons express neuron-specific markers
Immunofluorescent staining on day 11 post-revival demonstrates similar homogenous expression of pan-neuronal proteins MAP2 and TUBB3 (upper panel) and glutamatergic neuron-specific transporter VGLUT2 (lower panel) in CRISPRko-Ready ioGlutamatergic Neurons compared to ioGlutamatergic Neurons (io1001). 100X magnification (upper panel). 200x magnification (lower panel).

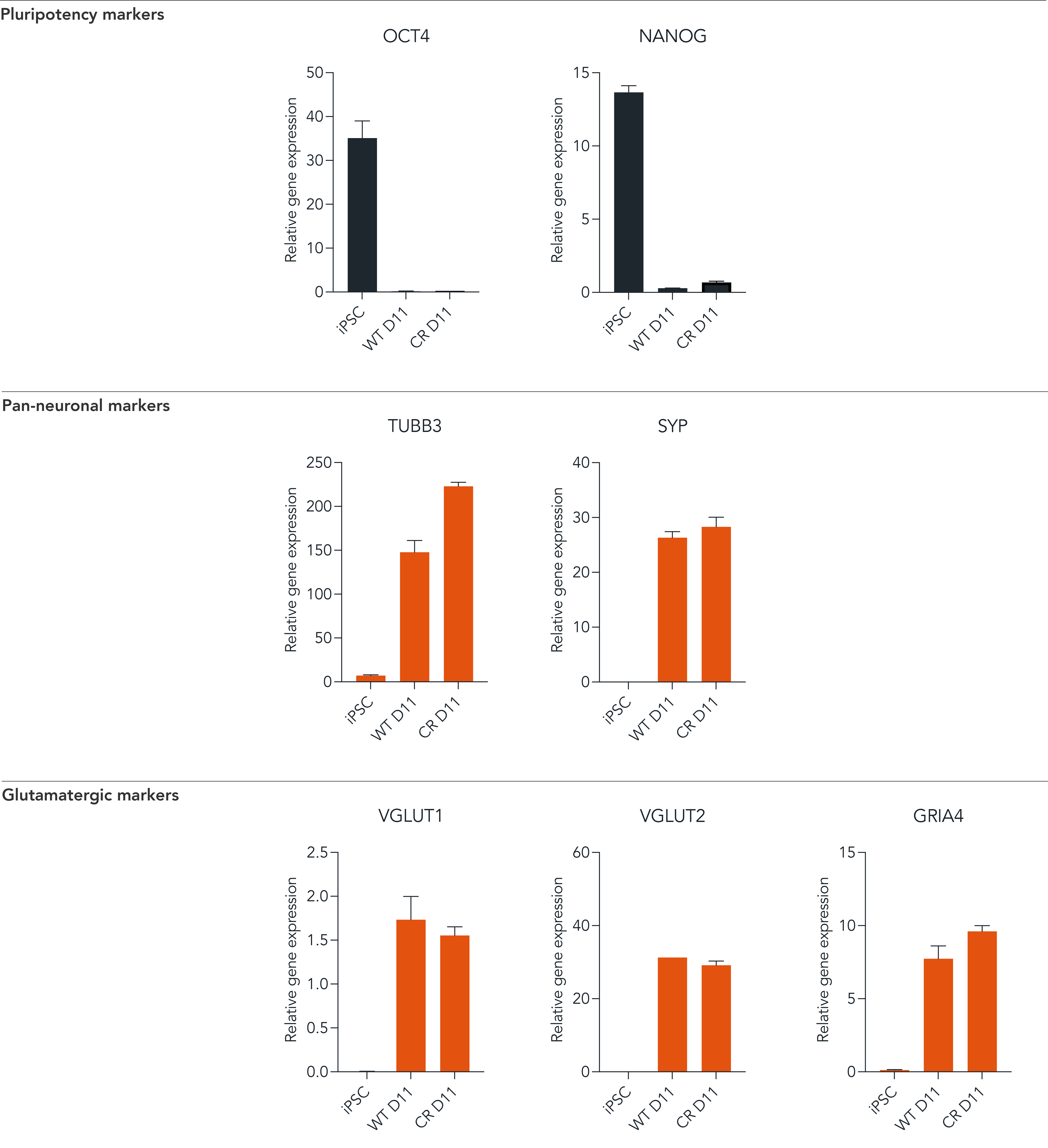
CRISPRko-Ready ioGlutamatergic Neurons demonstrate gene expression of neuronal-specific and glutamatergic-specific markers following deterministic cell programming
Gene expression analysis at day 11 demonstrates that CRISPRko-Ready ioGlutamatergic Neurons (CR) and ioGlutamatergic Neurons (WT) lack the expression of pluripotency markers (NANOG and OCT4). In contrast, they robustly express pan-neuronal (TUBB3 and SYP) and glutamatergic-specific (VGLUT1 and VGLUT2) markers, and the glutamate receptor GRIA4. Gene expression levels were assessed by RT-qPCR. Data normalised to HMBS; cDNA samples of the parental human iPSC line (iPSC) were included as reference; n=3 replicates.

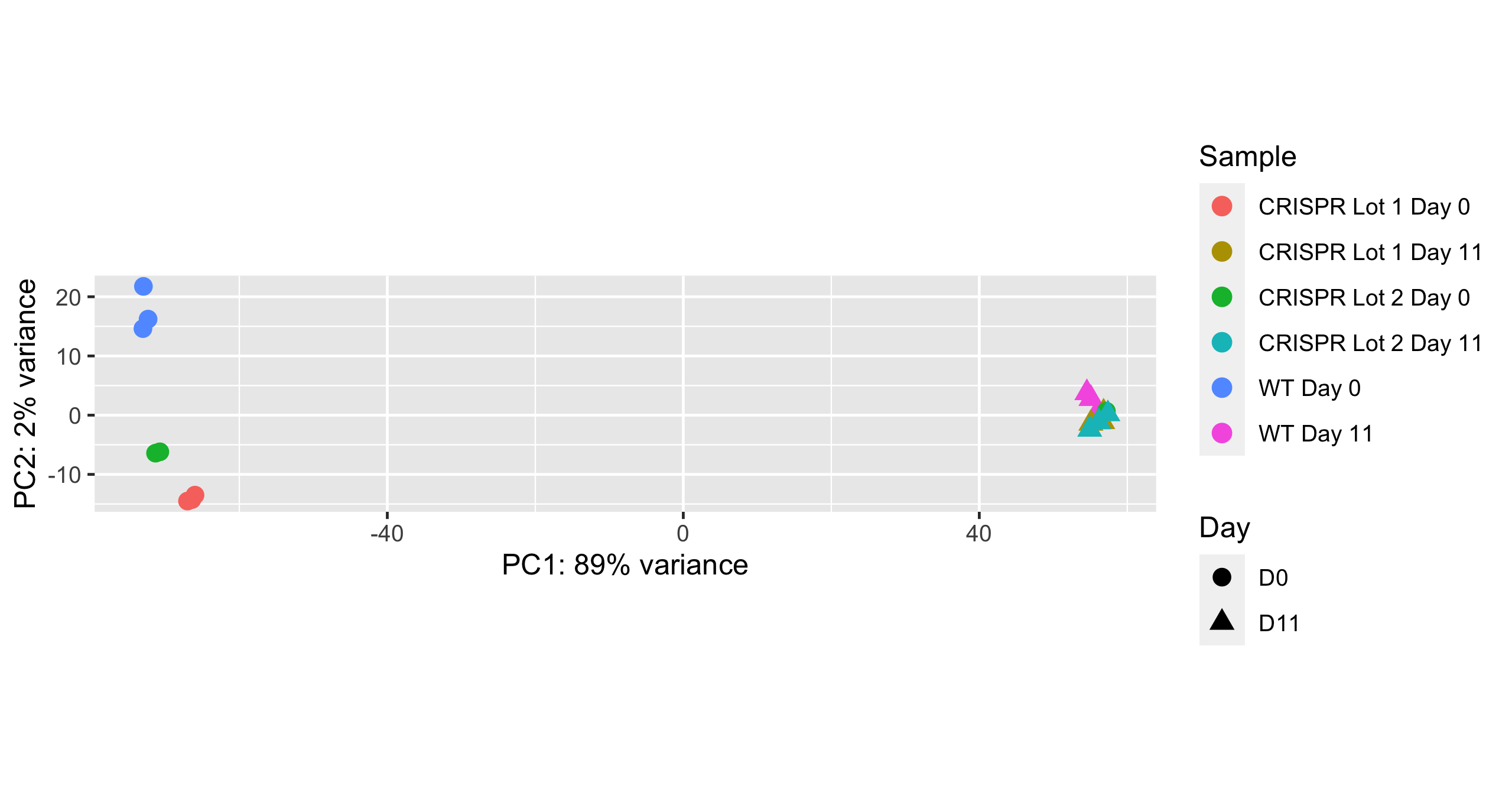
Whole transcriptome analysis demonstrates equivalent expression profiles between CRISPRko-Ready ioGlutamatergic Neurons and wild-type ioGlutamatergic Neurons
Bulk RNA sequencing analysis was performed on two independent lots of CRISPRko-Ready ioGlutamatergic Neurons (CRISPR) and one lot of wild-type ioGlutamatergic Neurons (WT) at day 0 and day 11 of the protocol. Principal component analysis represents the variance in gene expression between CRISPRko-Ready ioGlutamatergic Neurons and ioGlutamatergic Neurons and shows equivalent expression profiles between these cells. Shapes represent the day the samples from which data was obtained and colours represent the cell type and lot.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.


.png?width=1715&height=651&name=ioGlutamatergic_neurons-CRISPR-Morphology%20(1).png)
%20compressed.png?width=1715&height=1221&name=ioGlutamatergic_neurons-CRISPR-ICC%20(1)%20compressed.png)


_MAP2(R)_DAPI(B)_%20comp.jpg?width=604&name=a-HTT50CAGWT_Overlay__TUBB3(G)_MAP2(R)_DAPI(B)_%20comp.jpg)
Hoescht(blue)TUBB3(blue)_day4.jpg?width=604&name=bit.bio_ioGlutamatergic%20Neurons_60xMAP2(red)Hoescht(blue)TUBB3(blue)_day4.jpg)
 Emmanouil Metzakopian
Emmanouil Metzakopian




