














cat no | io1095
CRISPRko-Ready ioOligodendrocyte-like cells
Human iPSC-derived oligodendrocyte-like cells expressing Cas9
- Cryopreserved human iPSC-derived cells powered by opti-ox, that are ready for experiments in days
- Ideal for functional genomics, stably expressing Cas9 for gene knockouts and CRISPR screens
- Resemble a pre-myelinating oligodendrocyte expressing MBP marker by day 8

Human iPSC-derived oligodendrocyte-like cells expressing Cas9

Flow cytometry analysis reveals high knockout efficiency of beta-2 microglobulin (B2M) by lentiviral transduction
Flow cytometry analysis of B2M protein expression in CRISPRko-Ready ioOligodendrocyte-like cells after delivery of a guide RNA (gRNA) targeting B2M. gRNAs were introduced into the cells at day 2 post-thaw using lentiviral transduction and gene knockout was assessed 6 days post guide delivery. B2M was selected as a proof-of-concept target because it is widely expressed in the cells.
(A) Lentiviral transduction on day 2 with gRNA targeting B2M; approximately 52% of cells received a B2M gRNA, as measured by GFP expression.
(B) Functionality of Cas9 upon B2M gRNA delivery is confirmed, as demonstrated by the histograms showing loss of B2M protein expression in 95% of cells (orange) compared to the cells that received a non-targeting (NT) gRNA (grey); cells gated on the GFP+ population.

Go from seeding to knockout to readout in days

A pooled CRISPR screen in ioOligodendrocyte-like cells highlights genes involved in myelination
A pooled CRISPRko screen was performed in CRISPRko-Ready ioOligodendrocyte-like cells to identify positive and negative regulators of myelination. A total of 160 candidate genes with four sgRNAs per gene were selected for the whole-transcriptome analysis (WTA) screen. ioOligodendrocyte-like cells were transduced 2 days post-thawing and single-cell gene expression analysis was performed 5 days post transduction.
(A) Reliable, high-quality CRISPR screen results Quality control of the CRISPRko screen and sequencing metrics showed high data quality across two channels, including high sequencing saturation (>0.4) and robust median reads, UMI counts, and guide UMI counts per cell.
(B) Representation of gene perturbations in a pooled CRISPRko screen in ioOligodendrocyte-like cells Distribution of the number of cells per gene perturbation in the pooled CRISPRko screen, showing that most perturbations are represented by more than 100 cells, ensuring robust coverage per target gene.
(C) A pooled CRISPR screen in ioOligodendrocyte-like cells pinpoints the drivers and brakes of myelination Heatmap shows the effect of gene knockouts in ioOligodendrocyte-like cells on four core myelination marker genes.

CRISPRko-Ready ioOligodendrocyte-like cells express typical oligodendroglial-specific markers
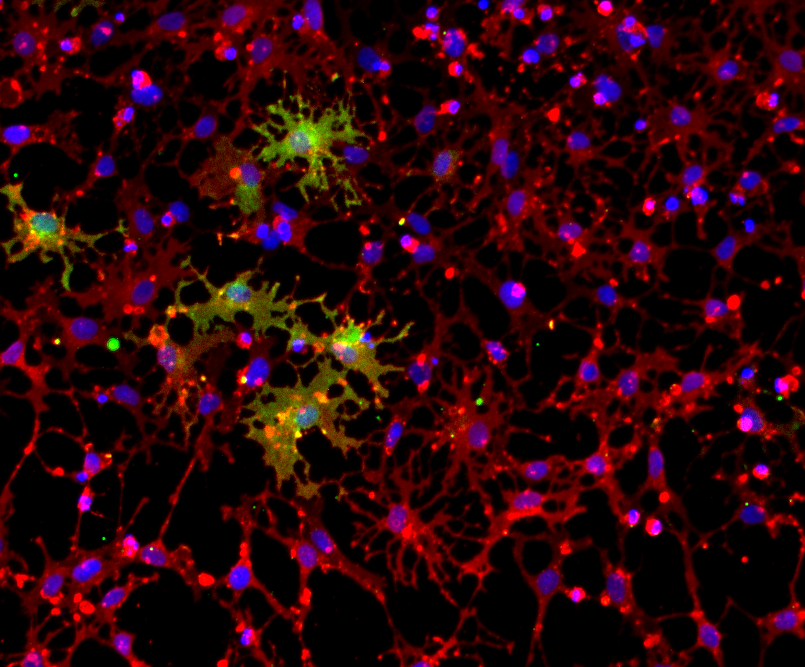
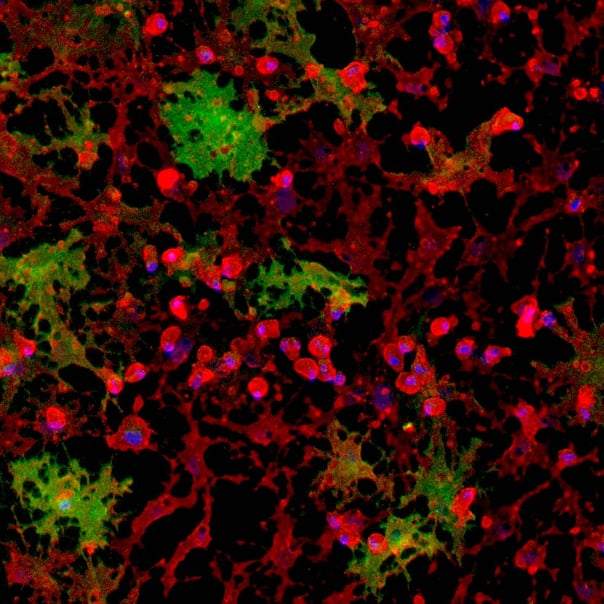
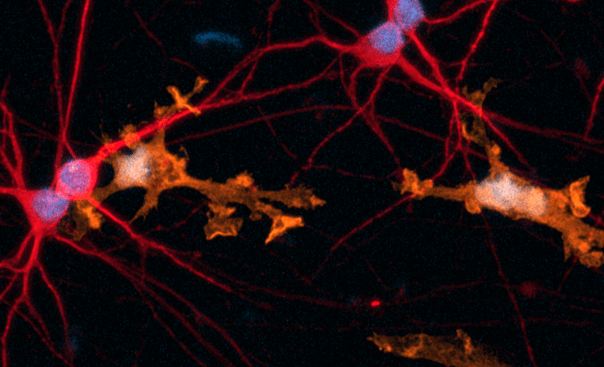
Immunofluorescent staining of the cells at day 1 (upper panel) and day 8 (lower panel) post-revival. Marker expression is highly similar between CRISPRko-Ready and wild-type ioOligodendrocyte-like cells. At day 1, the cells are positive for the oligodendrocyte-specific marker O4 (red), and the DAPI counterstain (blue). At day 8, cells show an increased complexity and are positive for O4 (red), the myelin basic protein (MBP) (green), and the DAPI counterstain (blue). Scale bar: 100 μm.

Whole transcriptome analysis demonstrates equivalent expression profiles between CRISPRko-Ready and wild-type ioOligodendrocyte-like cells
Bulk RNA-sequencing analysis was performed on CRISPRko-Ready and wild-type ioOligodendrocyte-like cells at day 1 and day 8. Principal component analysis (PCA) represents the variance in gene expression between the CRISPRko-Ready and wild-type ioOligodendrocyte-like cells. Differential expression analysis reveals no statistically significant differentially expressed (DE) genes between the two products, based on a log fold change threshold of 0.5 and an s-value cutoff of 0.01.
This analysis shows clustering of both products, demonstrating equivalent expression profiles at each given timepoint. Colours represent different samples; shapes represent different timepoints; n=3 replicates.
Expression levels for specific genes of interest can be requested by contacting our team at technical@bit.bio.

Key oligodendroglial genes are expressed by CRISPRko-Ready and wild-type ioOligodendrocyte-like cells
Gene expression analysis demonstrates that CRISPRko-Ready ioOligodendrocyte-like cells (CRISPRko-Ready) and wild-type ioOligodendrocyte-like cells (wild-type) have similar marker expression at two timepoints.
Following deterministic programming, the cells downregulate expression of the pluripotency genes OCT4, whilst demonstrating robust expression of relevant oligodendroglial markers, including PDGFRA, PLP1, MBP, and MAG. Gene expression levels assessed by RT-qPCR, data expressed relative to the reference (housekeeping) gene, HMBS. Data represents day 1 and day 8 post-thaw samples; cDNA samples of the parental human iPSC line (iPSC) were included as reference; n=3 technical replicates.

Cells rapidly mature, transitioning into an oligodendrocyte-like morphology within 8 days
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.



Hoescht(blue)TUBB3(blue)_day4.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_60xMAP2(red)Hoescht(blue)TUBB3(blue)_day4.png)




Hoescht(blue)_day12v2.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_20xMAP2(red)Hoescht(blue)_day12v2.png)