













cat no | io1101
CRISPRko-Ready ioMotor Neurons
Human iPSC-derived motor neurons expressing Cas9 for rapid gene knockout generation
- Cryopreserved human iPSC-derived cells powered by opti-ox that are ready for experiments in days
- Neurons for functional genomics, stably expressing Cas9 for gene knockouts and CRISPR screens
-
Clump free, highly-pure motor neurons, that form functional neuronal networks in co-culture with astrocytes

Human iPSC-derived motor neurons expressing Cas9 for rapid gene knockout generation

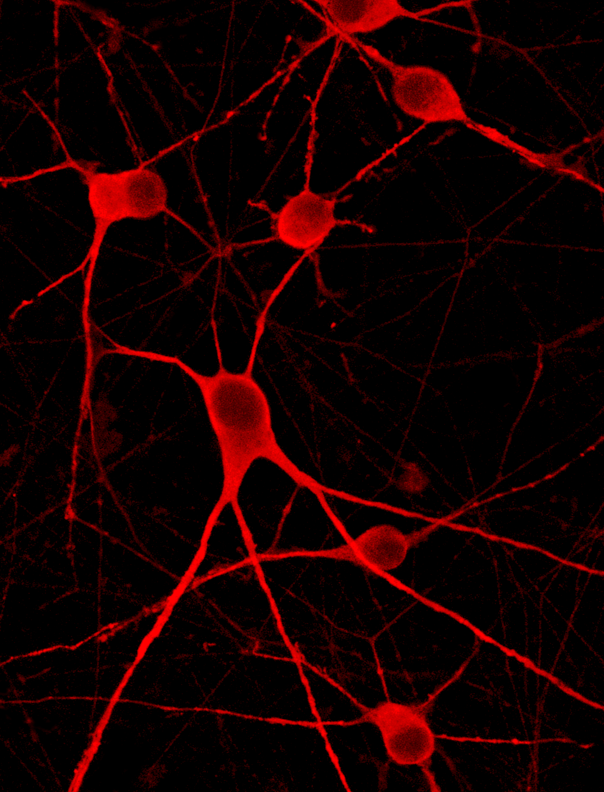
Immunofluorescence staining demonstrates high knockout efficiency of SOX11 by lentiviral transduction
Immunofluorescence staining of CRISPRko-Ready ioMotor Neurons demonstrates a highly efficient knockout of SOX11. The gRNA was delivered by lentiviral transduction on day 3 post-revival. Immunofluorescence staining of SOX11 was conducted five days post gRNA delivery (day 8 post-revival). A non-targeting gRNA was used as a control.

Immunofluorescence staining demonstrates high knockout efficiency of SOX11 by lentiviral transduction
Immunofluorescence quantification demonstrates >80% knockout efficiency of SOX11.

Flow cytometry analysis demonstrates high knockout efficiency CD63 by lentiviral transduction
Flow cytometry analysis of CD63 protein expression in CRISPRko-Ready ioMotor Neurons, after delivery of gRNA targeting CD63. gRNAs were introduced into the cells at day 3 post-thaw using lentiviral transduction. After 8 days of culture following guide delivery, CD63 gene knockout efficiency was assessed by flow cytometry analysis.
(C) Lentiviral transduction with gRNA targeting CD63: 53% of cells received a CD63 gRNA, as measured by GFP expression. A high knockout efficiency of 80% was achieved in these GFP+ cells (D) compared to (B) the non-targeting gRNA population.

CRISPRko-Ready ioMotor Neurons form structural neuronal networks from day 4
CRISPRko-Ready ioMotor Neurons mature rapidly, show motor neuron morphology and form structural neuronal networks over 11 days, highly similar to wild-type ioMotor Neurons (io1027). From Day 4 onwards cells begin to exhibit classical neuronal morphology with clear outgrowth, no signs of cell clustering or clumping. Day 1 to 11 post-thawing.

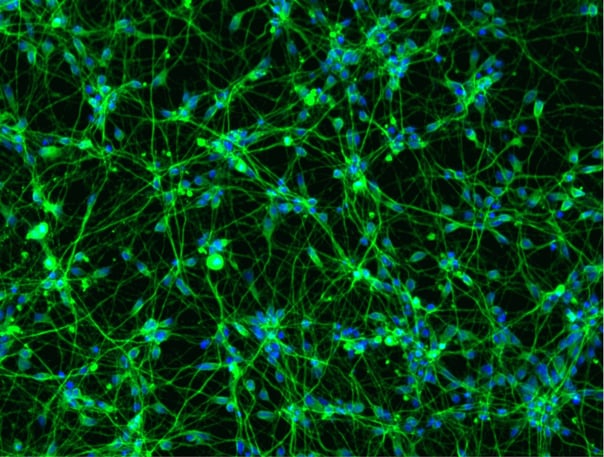
CRISPRko-Ready ioMotor Neurons express neuron-specific markers
Immunofluorescence staining at day 11 demonstrates that CRISPRko-Ready ioMotor Neurons and wild type ioMotor Neurons show comparable expression of key markers – indicating Cas9 expression has not affected cell programming. Both cells express pan-neuronal markers MAP2 and TUBB3, the cholinergic markers ChAT and VAChT and the motor neuron-specific markers MNX1 and ISL1/2.

CRISPRko-Ready ioMotor Neurons demonstrate gene expression of neuronal-specific and motor neuron markers following deterministic cell programming
Gene expression analysis at day 11 demonstrates that CRISPRko-Ready ioMotor Neurons (CR) and ioMotor Neurons (WT) lack the expression of pluripotency markers (NANOG and OCT4). In contrast, they robustly express pan-neuronal (TUBB3, MAP2) and cholinergic (CHAT, VACHT) markers, and the motor neuron specific markers (ISL2, HB9). Gene expression levels were assessed by RT-qPCR. Data normalised to HMBS; cDNA samples of the parental human iPSC line (iPSC) were included as reference; n=3 replicates.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.




Hoescht(blue)TUBB3(blue)_day4.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_60xMAP2(red)Hoescht(blue)TUBB3(blue)_day4.png)
 Emmanouil Metzakopian
Emmanouil Metzakopian




