cat no | io1019
ioSkeletal Myocytes DMD Exon 52 Deletion
Human iPSC-derived Duchenne muscular dystrophy model
-
Cryopreserved human iPSC-derived cells powered by opti-ox that are ready for experiments in days
-
Study Duchenne muscular dystrophy in a human in vitro model engineered with a DMD exon 52 deletion
-
Deletion results in weaker contraction and fatigue; dystrophin restored by ASO-mediated exon skipping

Human iPSC-derived Duchenne muscular dystrophy model

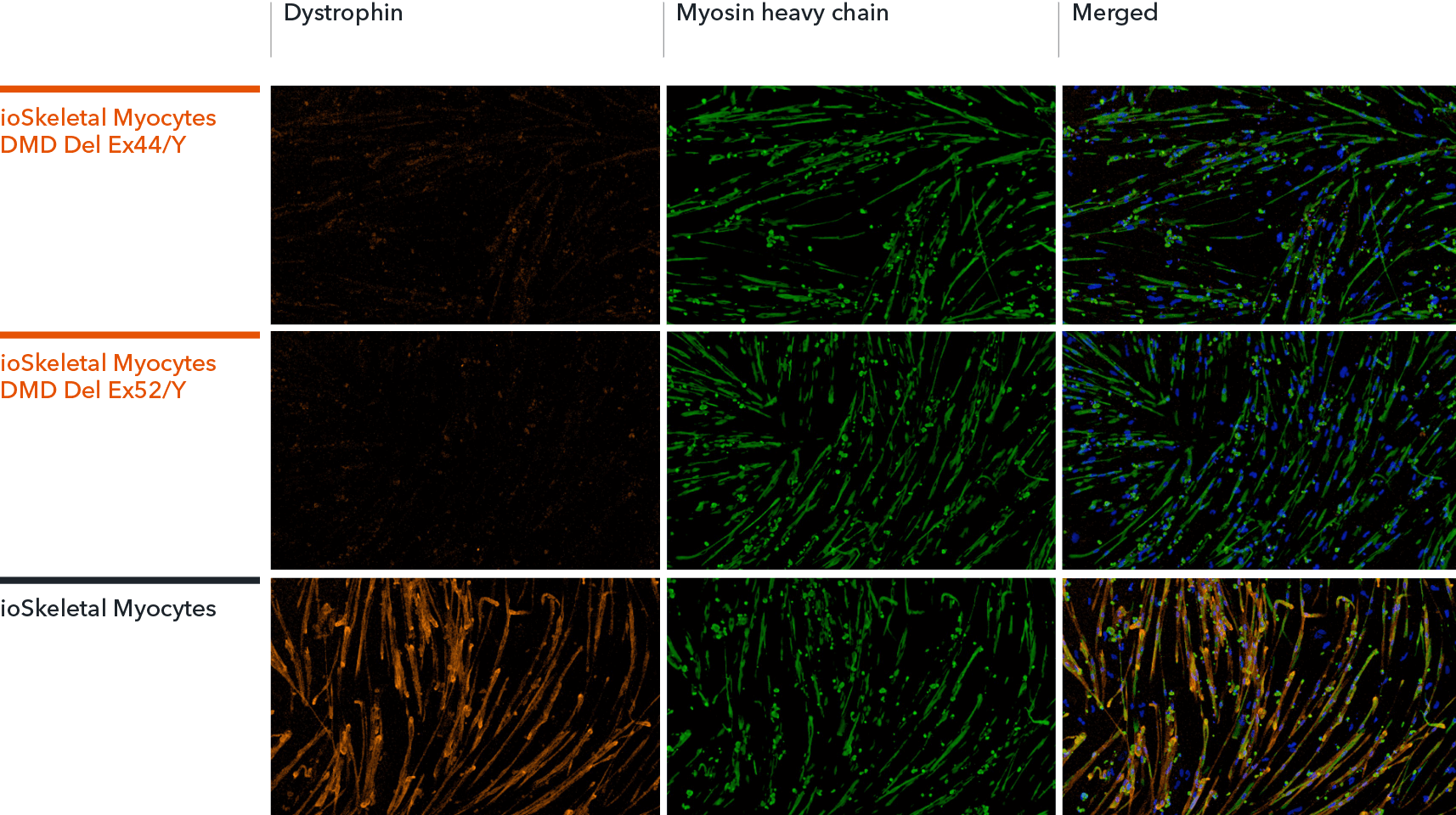
ioSkeletal Myocytes DMD Exon Deletion disease model cells show absence of Dystrophin protein by immunocytochemistry, demonstrating a Duchenne muscular dystrophy phenotype
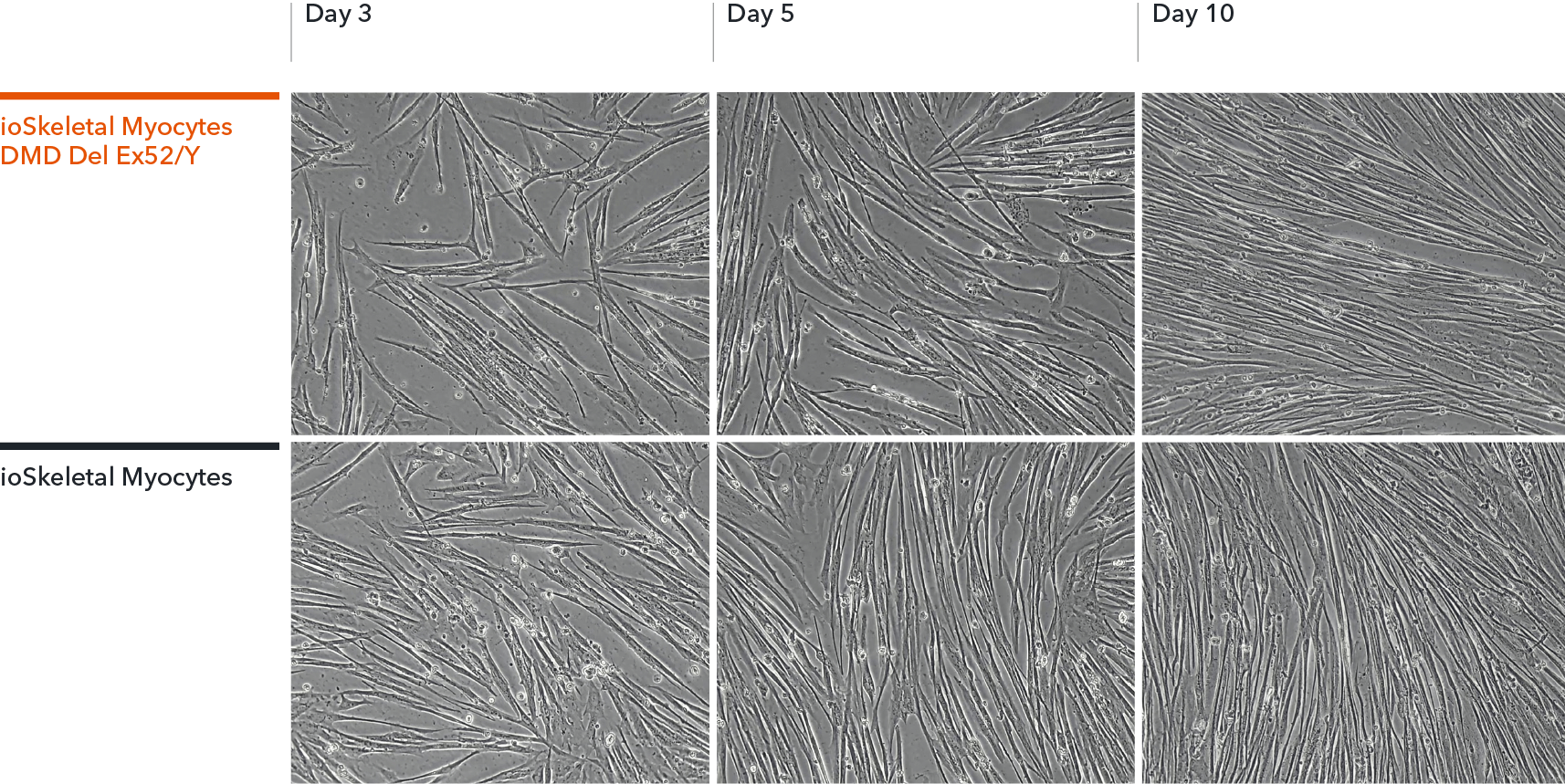
ioSkeletal Myocytes DMD Exon 44 Deletion and DMD Exon 52 Deletion disease model cells, and ioSkeletal Myocytes wild type isogenic control, were cultured in 96-well plates at a density of 32,000 cells per well, according to the user manual. Immunocytochemistry staining for Dystrophin and Myosin Heavy Chain (MHC) was carried out at day 10 post-revival. The data show robust expression of MHC, but no expression of Dystrophin in the ioSkeletal Myocytes DMD Del Ex44/Y or DMD Del Ex52/Y disease model cells compared to the wild-type control, demonstrating a Duchenne muscular dystrophy phenotype. Dystrophin was restored by ASO-mediated exon skipping. Data courtesy of Charles River Laboratories.

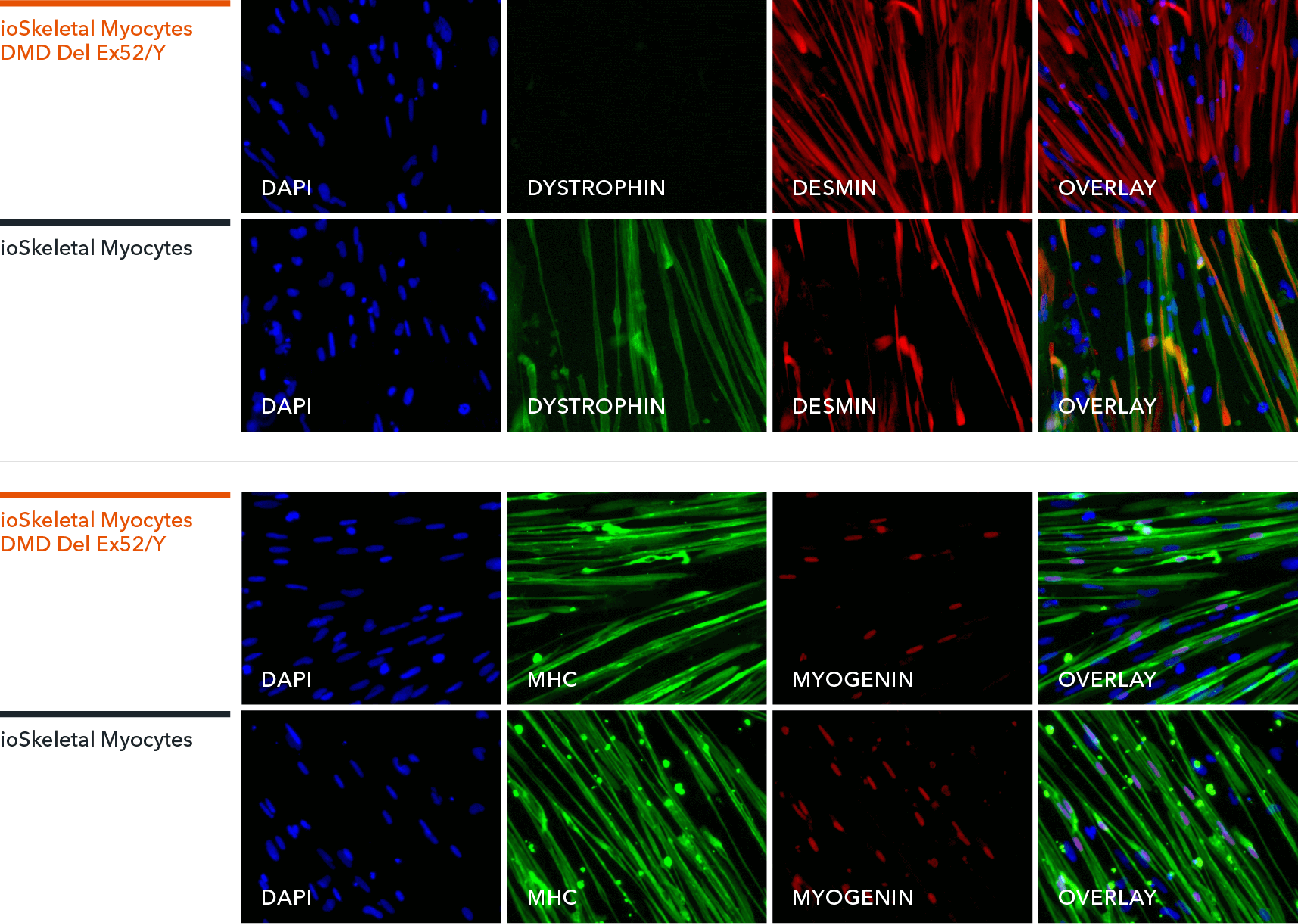
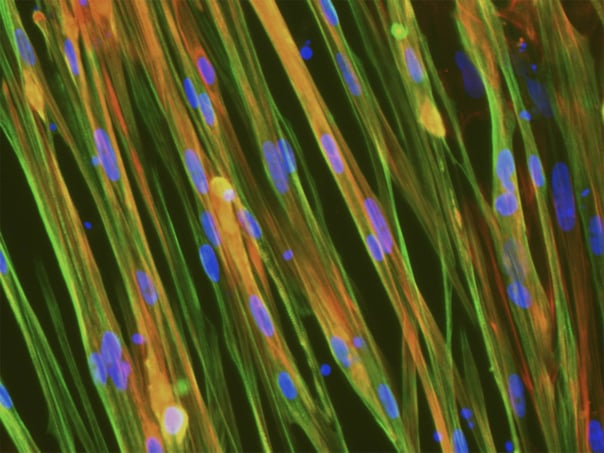
ioSkeletal Myocytes DMD Exon 52 Deletion disease model cells express skeletal muscle cell specific markers and lack expression of Dystrophin, demonstrating a Duchenne muscular dystrophy phenotype
Immunocytochemistry staining at day 10 post revival demonstrates robust expression of Desmin, a component of the contractile apparatus, and no expression of Dystrophin in the ioSkeletal Myocytes DMD Del Ex52/Y disease model cells, whereas ioSkeletal Myocytes, the wild type isogenic control, express both markers (upper panel). Robust expression of Myosin Heavy Chain (MHC) and the muscle transcription factor Myogenin is observed in ioSkeletal Myocytes DMD Del Ex52/Y and ioSkeletal Myocytes (lower panel). Anti-dystrophin antibody clone 2C6 (MANDYS106).

ioSkeletal Myocytes DMD Exon 52 Deletion disease model cells demonstrate classical skeletal myocyte morphology

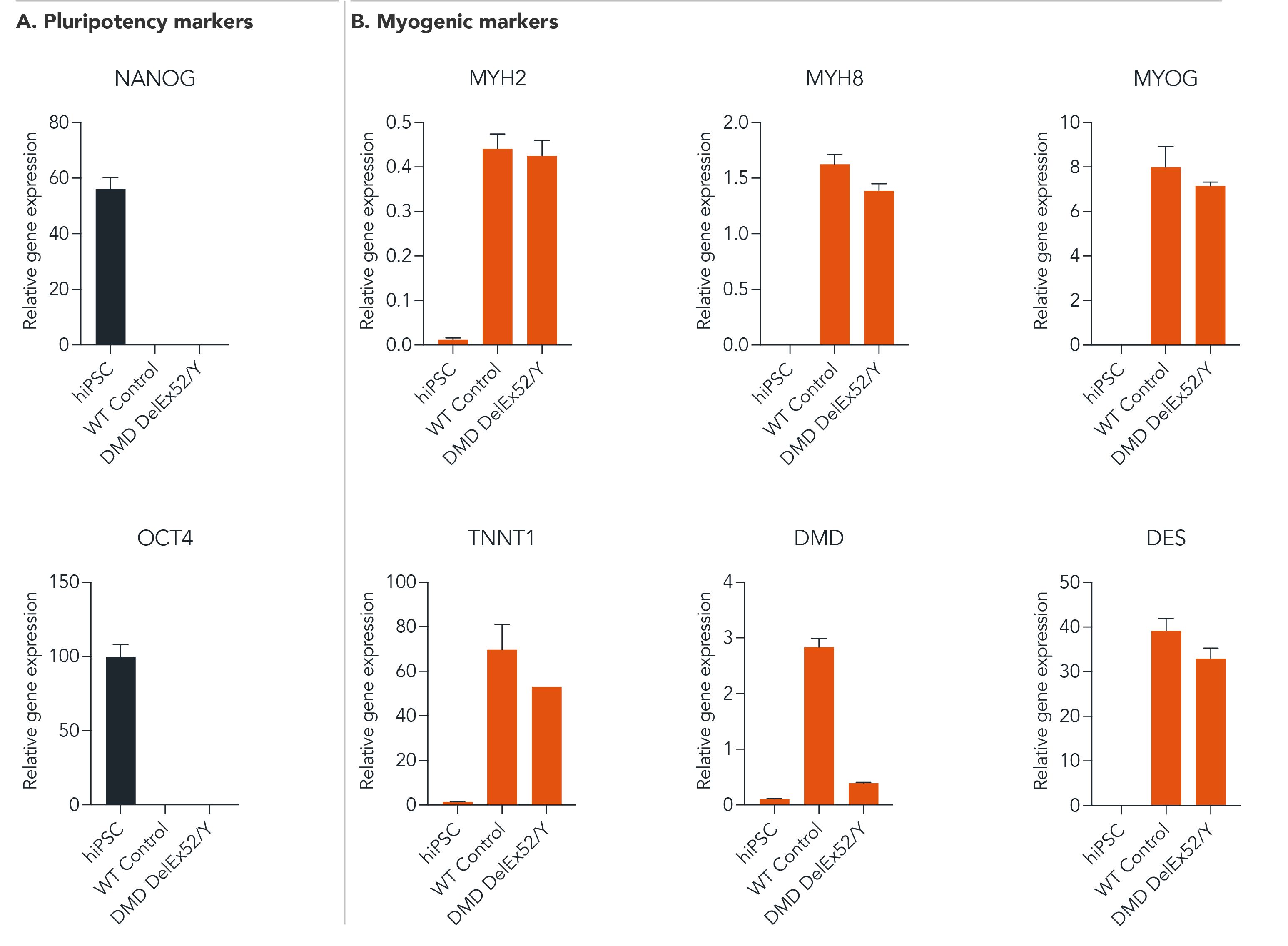
ioSkeletal Myocytes DMD Exon 52 Deletion disease model cells demonstrate gene expression of key myogenic markers following deterministic programming
Following reprogramming, ioSkeletal Myocytes DMD Exon 52 Deletion (DMD DelEx52/Y) and wild type ioSkeletal Myocytes (WT Control) downregulate expression of pluripotency genes (A), while demonstrating expected expression of key myogenic markers (B). Gene expression levels were assessed by RT-qPCR (data normalised to HMBS; cDNA samples of the parental human iPSC line (hiPSC) were included as reference). Data represents day 10 post-revival samples, (n=2 replicates).

Industry leading seeding density
ioSkeletal Myocytes DMD Del Ex52/Y are compatible with plates ranging from 6 to 384 wells and are available in two vial sizes, tailored to suit your experimental needs with minimal waste. The recommended seeding density is 100,000 cells/cm2.
One small vial can plate a minimum of 0.5 x 24-well plate, 0.75 x 96-well plate, or 1 x 384-well plate. One large vial can plate a minimum of 1 x 24-well plate, 1.5 x 96-well plates, or 2 x 384-well plates.

Dose-dependent ASO-mediated dystrophin restoration in ioSkeletal Myocytes DMD Exon 52 Deletion
An exon 51 skipping phosphorodiamidate morpholino oligomer (PMO) antisense oligonucleotide (ASO) was designed based on Heemskerk et al., 2009 J Gene Med.
Efficacy was tested in ioSkeletal Myocytes DMD Exon 52 Deletion. A control non-skipping ASO (CTR PMO) and the Exon 51 skipping ASO (Ex51 PMO) were delivered by gymnosis on day 7. The Ex51 PMO ASO restored dystrophin protein in a dose-dependent manner.
Data courtesy of Mitchell Han and Marieke Aarts, formerly at Bi/ond Solutions B.V.

DMD Exon Deletion 3D muscle microtissues show weaker contraction and fatigue
Wild-type (WT) and DMD exon deletion disease model ioSkeletal Myocytes were cultured in 3D on the MUSbit microchip (Bi/ond), which includes pillars designed for anchoring muscle cell bundles. Muscle microtissues developed over 14 days.
When compared to the genetically matched control (WT) at day 14, the DMD Exon 44 Deletion and Exon 52 Deletion models showed weaker contraction upon twitch and tetanus stimuli (A), and fatigue under sustained stimulation (B).
The 3D muscle microtissues provide a relevant model to study how exon deletions in the dystrophin gene affect muscle contraction.
Data courtesy of M. Han and M. Aarts, formerly at Bi/ond Solutions BV.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.


-1%20(1).png?width=604&name=horizontal_ioSkeletal-Myocytes-Muscle-Bundle-Biond-ICC-SAA-Actin%20(1)-1%20(1).png)
.png?width=604&name=ISSCR24-DMD-Exon44-3D-muscle-bundle%20(1).png)