cat no | io1085
ioGABAergic Neurons APP V717I/WT
Human iPSC-derived Alzheimer's disease model
-
Cryopreserved human iPSC-derived cells powered by opti-ox, that are ready for experiments in days
-
Engineered to enable investigations into the effect of the mutant APP protein on Alzheimer's disease
-
Disease-related phenotype demonstrated by increased amyloid beta production vs. wild-type control

Human iPSC-derived Alzheimer's disease model

Increased ratio of A𝛽42:40 seen in ioGABAergic Neurons APP V717I (London), as observed in Alzheimer’s disease
ioGABAergic Neurons APP V717I/WT disease model (HET) cells show a trend of increased production of A𝛽38 peptides and A𝛽42 (involved in the amyloidogenic pathway), with minimal difference seen for A𝛽40 (A). This results in a significantly increased ratio of A𝛽42:40 and no change in the A𝛽42:38 ratio (B).
-
ioGABAergic neurons (WT, CL54, CL65, CL59, CL70) were seeded at 150,000 cells/cm2 in 24 well plates and cultured for 30 days according to the user manual.
-
Supernatant was collected at days 10, 20, and 30 to quantify levels of A𝛽38, A𝛽40, A𝛽42 peptides using the V-PLEX Aβ Peptide Panel ELISA kit (MSD K15200E-1).
-
Concentrations of A𝛽38, A𝛽40, A𝛽42 were normalised to the calculated total number of cells per well.
-
Data were obtained from two independent experiments and are shown as mean ± SEM. Data were analysed statistically (at days 20 and 30) using one-way ANOVA with Tukey’s post-hoc analysis. * p<0.05 ** p<0.01 ***p<0.001 **** p<0.0001

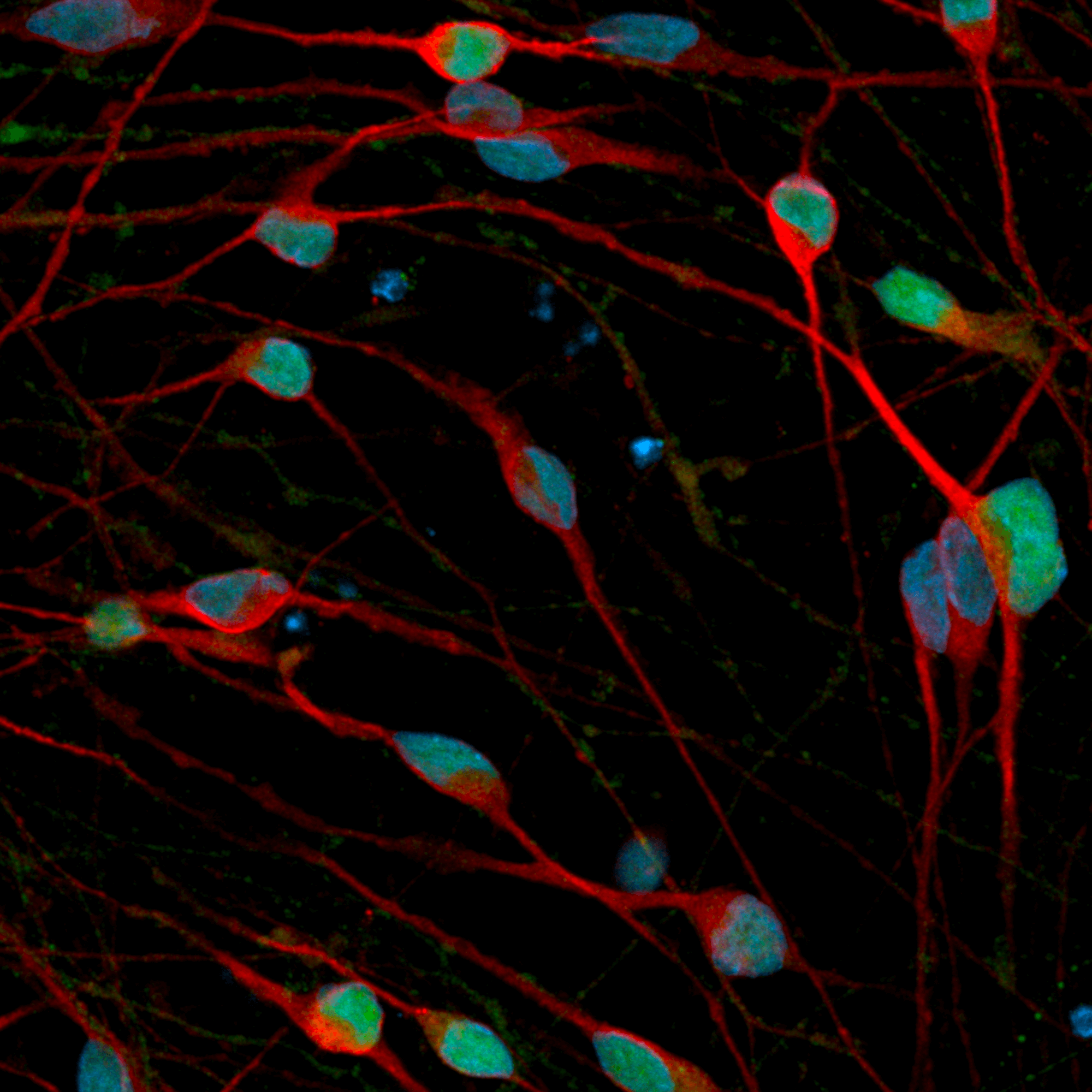
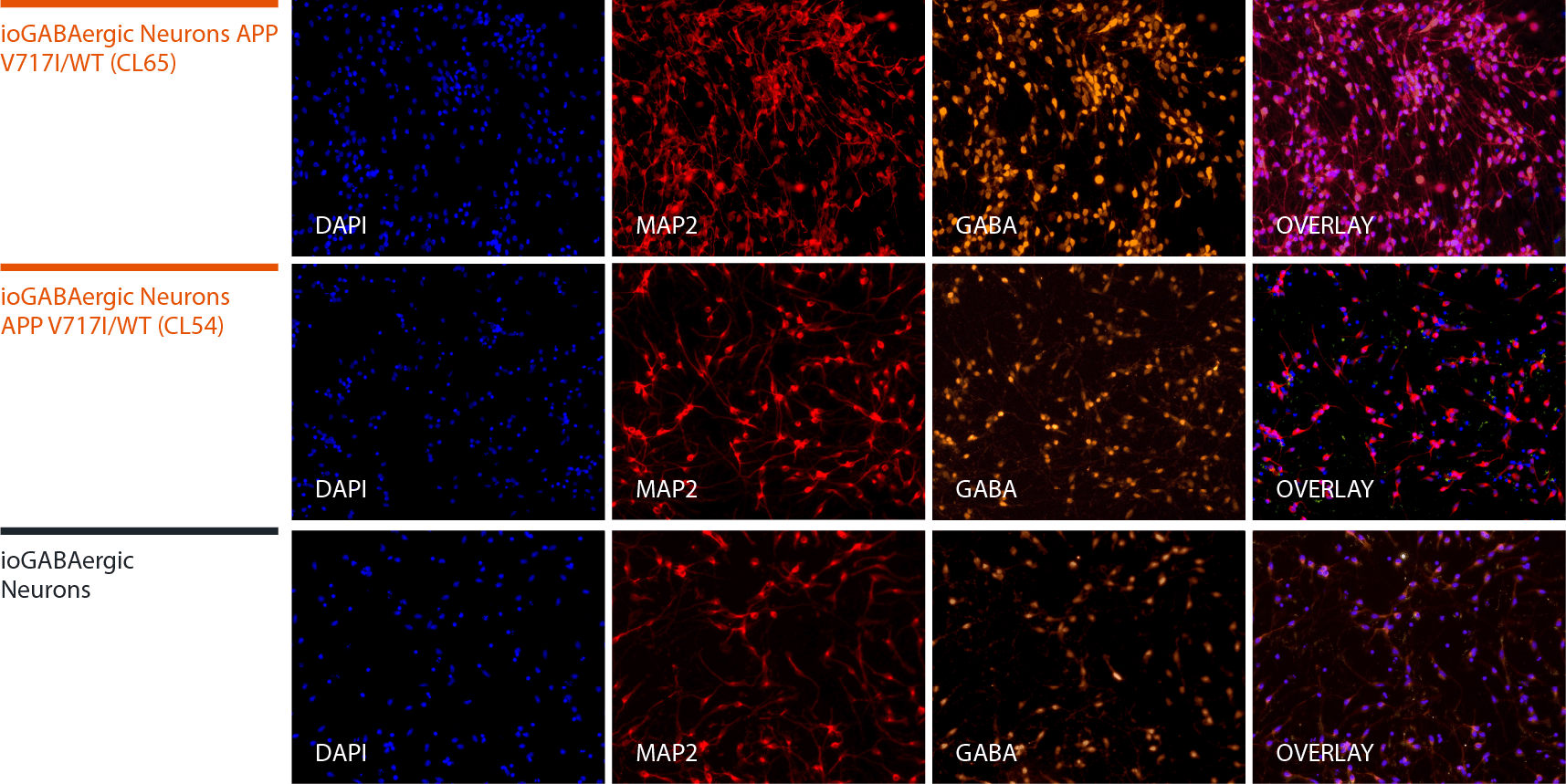
Disease model cells express key GABAergic neuron-specific markers comparably to the isogenic control
Immunofluorescent staining on day 12 post-revival demonstrates similar homogenous expression of the pan-neuronal marker, MAP2 and GABAergic neuron-specific marker, GABA in both disease model clones compared to the wild-type (WT) isogenic control. 100X magnification.

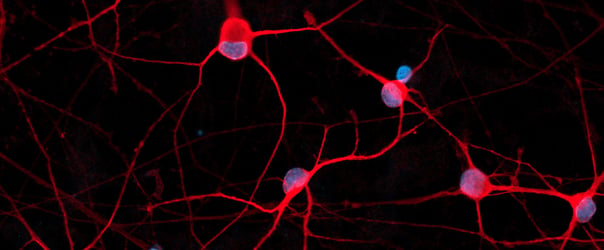
Disease model cells form structural neuronal networks by day 12

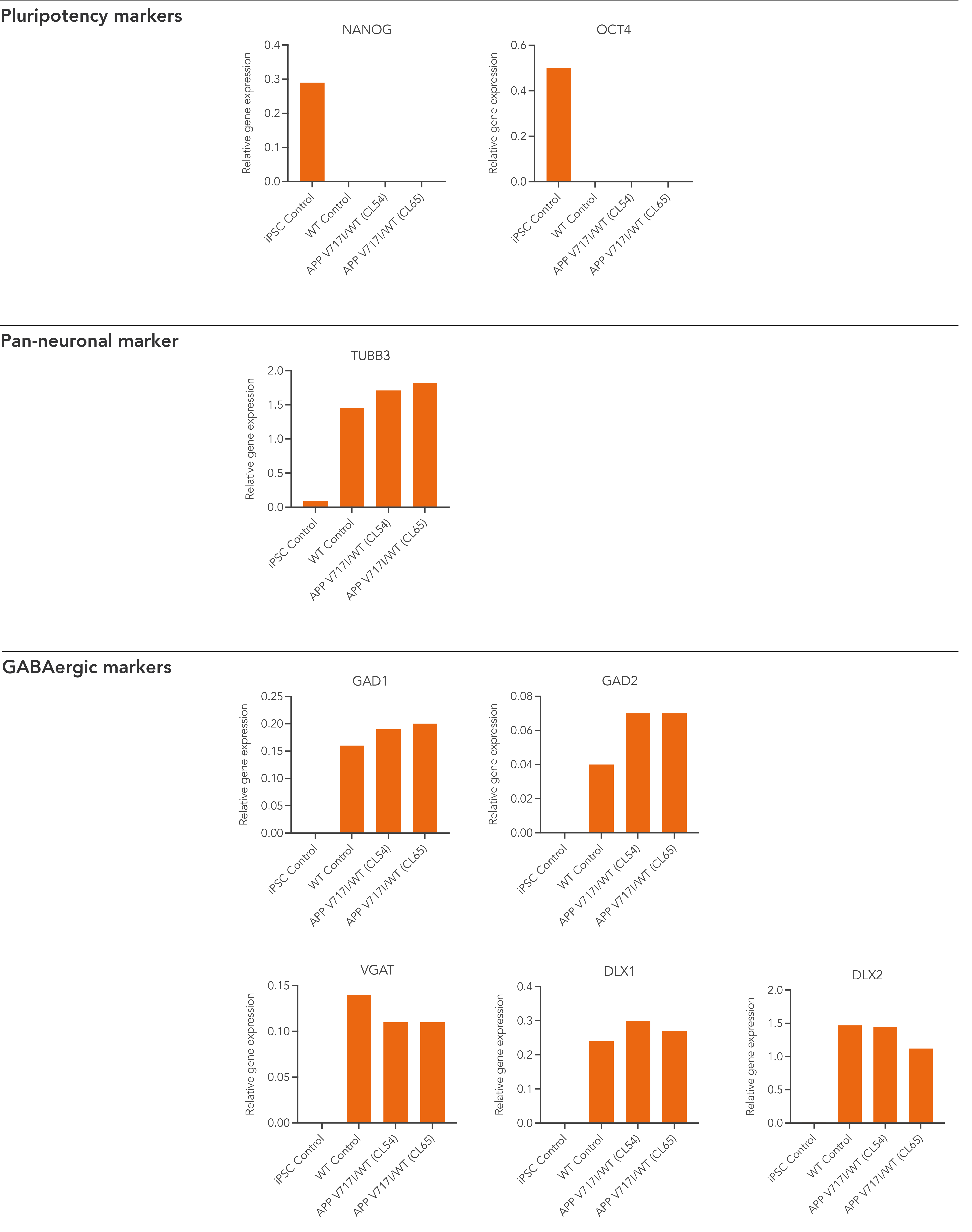
Disease model cells demonstrate gene expression of neuronal and GABAergic-specific markers following deterministic programming
Gene expression analysis demonstrates that both disease model clones and the WT isogenic control lack the expression of pluripotency markers (NANOG and OCT4) at day 12 post-thaw, whilst robustly expressing pan-neuronal (TUBB3) and GABAergic-specific markers, GAD1, GAD2, VGAT, DLX1, and DLX2. Gene expression levels were assessed by RT-qPCR (data expressed relative to the parental hiPSC control (iPSC Control), normalised to GAPDH). Data represents day 12 post-revival samples.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.



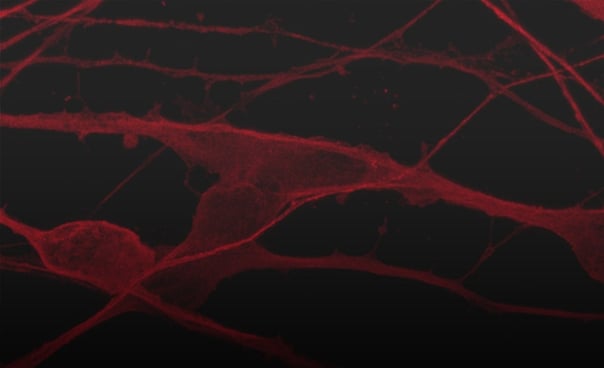
_MAP2(R)_Tubb3(B)_Hoechst(B)_20x_merge-comp.jpg?width=604&name=Colour%20webinar%20with%20it-bio%20ioGlutamatergic%20Neurons_VGLUT2(G)_MAP2(R)_Tubb3(B)_Hoechst(B)_20x_merge-comp.jpg)
Hoescht(blue)_day12v2.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_20xMAP2(red)Hoescht(blue)_day12v2.png)