28.10.2025 | Published by bit.bio
Despite decades of innovation, attrition rates in drug discovery remain unacceptably high: fewer than 1 in 10 candidates entering clinical trials reach patients, and central nervous system (CNS) programs fail up to 90% of the time1,2. The root cause lies in the translational gap: current models fail to reliably predict human outcomes. Closing this gap requires defined, human-relevant in vitro systems that can support confident decision-making across the discovery workflow.
Human iPSC cell culture is helping to answer this need by providing a foundation for robust phenotypic screening across target ID, assay development, hit-to-lead, lead optimisation, and safety profiling.
The need for better cell-based assays
Cell-based assays in drug discovery have long relied on traditional models. While these systems remain workhorses of preclinical drug discovery, their limitations are now clearer than ever:
- Immortalised lines are robust and scalable, but their lack of phenotypic fidelity means signals often don’t translate, creating false positives and wasted effort downstream.
- Animal primary cells capture some physiology, but their variability, and species differences make it hard to generate reliable, human-relevant data at scale.
- Conventional iPSC-derived models offer human-derived cells, but poor purity and batch variability add noise, making it difficult to compare results across experiments or sites.
Commonly used in vitro models
For a deeper comparison of commonly used models — including the strengths and limitations of primary cells and immortalised lines — see our earlier blog: Primary Cells vs. Cell Lines: Is It Not Time for Something Better? Read the blog
The result is a lack of reproducibility in phenotypic screening, where subtle readouts must be compared across replicates, time points, and sites. This lack of reproducibility has not gone unnoticed. Regulators and funders are responding. The FDA has published a roadmap to reducing animal testing in preclinical safety studies3 and announced a 2025 plan to phase out animal testing requirements for monoclonal antibodies4. The NIH has also prioritised new funding for human-based research as the future of preclinical discovery5.
Human iPSC-derived models: progress and limitations
Human iPSC culture is already proving valuable in cell-based drug discovery. Unlike immortalised lines or animal-derived primary cells, they provide access to diverse human cell types, including neurons, cardiomyocytes, and hepatocytes. This makes them attractive for early-stage research, where capturing human-relevant biology is critical.
Across the discovery workflow, iPSC-derived cells are being applied in diverse ways, such as:
- Target identification and validation
Human iPSC-derived cells add value at the initial stages of drug discovery by offering a representation of human biology and disease in vitro, supporting accurate drug development from start to finish. iPSC-derived cells are compatible with genome editing approaches such as CRISPR, which enable pathway analysis and functional validation of targets directly in a human cellular context6. High-content screening (HCS) has also been widely used with iPSC-derived cells, combining automated imaging and quantitative analysis to measure reporter signals, morphology, and subcellular localisation7. - Assay development and optimisation
Using human iPSC-derived cells allows assays to be built around human-relevant pathways, ion channels, receptors, and transcriptional programs. Consequently, assay parameters (e.g., compound dosing, time course, endpoints) can be optimised based on human kinetics and signalling. For example, iPSC-derived neurons and cardiomyocytes are used in electrophysiology and calcium flux assays to measure excitability, ion channel function, and network activity8. These models also support optimisation of HCS protocols and real-time impedance assays to monitor morphology and proliferation7. - Hit-to-lead screening
During the hit-to-lead stage, iPSC-derived cells add human relevance for establishing structure–activity relationships (SAR) between chemical structure and biological activity. iPSC-derived hepatocytes are being introduced for use in metabolism and drug–drug interaction studies, including cytochrome P450 induction and inhibition9. As phenotypic and neuroinflammation assays evolve to improve physiological relevance, scientists are increasingly incorporating iPSC-derived neuronal and immune cells, often combined with viability or cytotoxicity readouts such as LDH release or apoptosis assays10. - Lead optimisation
iPSC-derived sensory neurons characterised via multi-electrode arrays (MEA) and stimulus responses are being used to model pain pathways and evaluate compound effects11,12. Many iPSC-derived cell types exhibit measurable physiological activity (e.g., neuronal firing), enabling optimisation for distinct functional readouts across targets and compound classes. - Safety and toxicology screens
iPSC-derived cardiomyocytes are widely used in the early stages of preclinical safety studies to assess pro-arrhythmic risk13. The CiPA initiative exemplifies this integration: over the past decade, iPSC-derived cardiomyocytes have been characterised across multiple sites and assay platforms using reference compounds, helping to standardise and enhance early-stage pro-arrhythmic risk assessment14. Hepatocyte models have also been benchmarked in drug-induced liver injury (DILI) studies, showing time- and dose-dependent toxicity consistent with known clinical outcomes15.
These examples highlight how iPSC-derived cells complement traditional systems, enabling scientists to generate human-relevant data earlier in discovery. However, most conventional iPSC-derived cells rely on directed differentiation protocols, which can be slow, technically demanding, and prone to variability—leading to batch-to-batch inconsistency that limits reproducibility and scalability in phenotypic screening16. Combined with persistently high clinical failure rates, these challenges highlight the urgent need for more consistent, human-relevant iPSC-derived cells1,2. Addressing these bottlenecks requires solutions that preserve human relevance while eliminating variability and scale constraints.
ioCells in cell-based drug discovery
The limitations of conventional iPSC differentiation can undermine otherwise powerful in vitro models. ioCells address these challenges through deterministic cell programming with opti-ox™, a gene targeting strategy that ensures every iPSC in a culture is programmed to the same defined cell identity.
ioCells are already being applied in key cell-based assays in drug discovery, across the workflow in areas where reproducibility and scalability are most critical:
- Target identification and validation
CRISPR-Ready ioCells are used in functional genomics studies, where reproducibility is essential for interpreting large-scale perturbation screens. For example, CRISPRko-Ready ioMicroglia engineered to express Cas9 have supported pooled CRISPR knockout experiments to identify regulators of immune activation pathways. - Assay development and optimisation
Assay reproducibility depends on consistent cell populations. ioCells, characterised for marker expression and function, have been applied in both monoculture and multi-cell culture systems. For example, ioGlutamatergic Neurons have been incorporated into 3D neuronal microtissues that retain functional stability across experiments. - Hit-to-lead screening
Human ioMicroglia have been used to study chemotaxis, cytokine release, and phagocytosis. In these assays, reproducibility across batches reduces variability, helping researchers distinguish between compound effects and noise. - Lead optimisation
ioCells have supported longitudinal electrophysiology and imaging studies, where consistent function over time is necessary for comparing candidate effects. In well characterised, defined and physiologically relevant disease models, such as ioGlutamatergic Neurons carrying ALS mutations, robust electrophysiological assays reproducible MEA support a more confident interpretation of drug responses. - Safety and toxicology screens
Human ioHepatocytes are being developed and applied to advance in vitro safety testing, providing defined and reproducible human models for assessing drug metabolism and hepatotoxicity. Their stable function and gene expression enable long-term assays for DILI, supporting consistent and translationally relevant results. Recent work highlights how ioHepatocytes are bridging discovery and toxicology research—accelerating the development of more predictive human liver models for safety assessment.
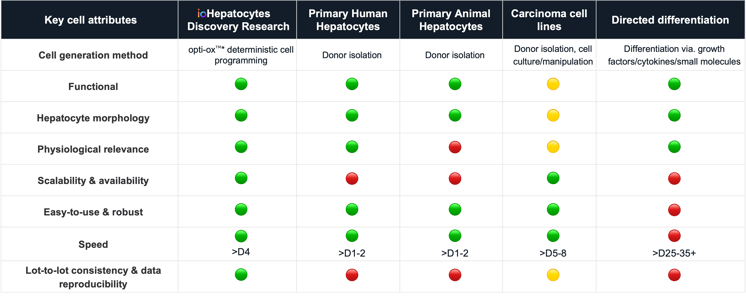
Overcoming the challenges of existing liver models | Comparing hepatocyte models highlights the need for scalable, reproducible human systems. As the liver is central to toxicity, introducing consistent and defined iPSC-derived ioHepatocytes earlier can improve safety insights and accelerate drug development timelines.
Key benefits of next-generation iPSC models
Unlike conventional directed differentiation, which can be long, complex, slow, and variable, opti-ox enables the rapid generation of consistent, defined human cell types. ioCells offer researchers control and confidence by providing consistent, specified inputs for discovery workflows.
Key benefits include:- Defined identity: every cell is programmed to the same fate, with <1% differential gene expression between lots.
- Consistency at scale: billions of consistently programmed cells can be generated in a single manufacturing run, ensuring supply for extended phenotypic screening campaigns.
- Ease of use: cells arrive cryopreserved and require a simple protocol using an open-source medium to get started.
- Assay versatility: ioCells support applications from functional genomics to complex co-culture electrophysiology.
Real-world impact
The advantages of reproducible, human-relevant cells are already being realised across preclinical drug discovery workflows:
CRO integration
Charles River Laboratories applied ioCells in neuroinflammation and demyelination assays, using complex co-culture models to improve translational predictivity in neurodegenerative research.
Disease modelling
ioGlutamatergic Neurons produced reproducible Huntington’s disease phenotypes, including mitochondrial dysfunction detectable by Seahorse assays—enabling more predictive therapeutic screens.
Functional characterisation
In collaboration with Axion Biosystems, ioCells were used in ALS disease models on MEA, revealing network-level deficits in TDP-43 mutant neurons compared to controls.
Improved reproducibility
Concept Life Sciences used ioMicroglia to replace rodent models, gaining a consistent, human-relevant system:
“ioMicroglia from bit.bio offered a solution by providing consistent and reproducible human iPSC-derived microglial cells.”
Elise Malavasi, PhD, Principal Scientist, Concept Life Sciences
View the application note
Download the SFN poster
These examples highlight how reproducibility and human relevance translate into tangible scientific progress, reinforcing the role of ioCells as cell-based assays in drug discovery.
The future of cell-based drug discovery
The challenge facing modern cell-based drug discovery is not simply choosing between immortalised lines, primary cells, or iPSC-derived models, but ensuring that any system can deliver reproducible, scalable, and human-relevant data that improves translational success.
Human iPSC-derived cells have already demonstrated their value across the workflow, from target identification to safety profiling. Yet conventional differentiation workflows remain variable, limiting their impact on large-scale discovery. Deterministically programmed ioCells address this gap by combining the human relevance of iPSC-derived systems with the reproducibility required for phenotypic screening at scale.
This direction is reinforced by the broader shift in the field: regulatory agencies and funders are explicitly prioritising human-based models. Initiatives such as the FDA’s Roadmap to Reducing Animal Testing3 and the NIH Complement-ARIE program17 show that reproducibility and human relevance are no longer optional, but expected.
As attrition rates remain high, the need for models that better predict human outcomes is urgent. ioCells represent one step toward that goal: not just an alternative to existing systems, but part of a new standard in cell-based drug discovery.
Ready to transform your phenotypic screening workflows?
ioCells provide reproducible, human-relevant cells that are ready for experiments within days. Their consistency reduces variability, enabling phenotypic screening campaigns that generate reliable, translational data.
References
- BIO IP, Advisors QL. Clinical Development Success Rates and Contributing Factors 2011–2020. BIO. https://www. bio. org/clinicaldevelopment-success-rates-and-contributing-factors-2011-2020. 2021.
- WCG Clinical. CNS Trial Failure Rates High As Need For New Drugs Grows. Clinical Leader. 2023.
- FDA, Roadmap to Reducing Animal Testing in Preclinical Safety Studies, available at https://www.fda.gov/media/186092/download?attachment.
- FDA News Release, “FDA Announces Plan to Phase Out Animal Testing Requirement for Monoclonal Antibodies and Other Drugs” (Apr. 10, 2025).
- NIH. Funding announcements to align with NIH initiative to prioritize human-based research. NIH Extramural Nexus. Published July 2025. Available at: https://grants.nih.gov/news-events/nih-extramural-nexus-news/2025/07/nih-funding-announcements-to-align-with-nih-initiative-to-prioritize-human-based-research. Accessed 23/09/2025.
- Bock C, Datlinger P, Chardon F, Coelho MA, Dong MB, Lawson KA, Lu T, et al. High-content CRISPR screening. Nat Rev Methods Primers. 2022;2:8. doi:10.1038/s43586-021-00096-x.
- Way GP, Sailem H, Shave S, Kasprowicz R, Carragher NO. Evolution and impact of high content imaging. Slas Discovery. 2023 Oct 1;28(7):292-305.
- Boutin ME, Strong CE, Van Hese B, Hu X, Itkin Z, Chen YC, LaCroix A, Gordon R, Guicherit O, Carromeu C, Kundu S. A multiparametric calcium signal screening platform using iPSC-derived cortical neural spheroids. SLAS Discovery. 2022 Jun 1;27(4):209-18.
- Corbett JL, Duncan SA. iPSC-derived hepatocytes as a platform for disease modeling and drug discovery. Frontiers in medicine. 2019 Nov 15;6:265.
- Stöberl N, Maguire E, Salis E, Shaw B, Hall-Roberts H. Human iPSC-derived glia models for the study of neuroinflammation. Journal of Neuroinflammation. 2023 Oct 10;20(1):231.
- Hiranuma M, Okuda Y, Fujii Y, Richard JP, Watanabe T. Characterization of human iPSC-derived sensory neurons and their functional assessment using multi electrode array. Scientific Reports. 2024 Mar 12;14(1):6011.
- Fofie CK, Granja-Vazquez R, Truong V, Walsh P, Price T, Biswas S, Dussor G, Pancrazio J, Kolber B. Profiling human iPSC-derived sensory neurons for analgesic drug screening using a multi-electrode array. Cell Reports Methods. 2025 May 19;5(5).
- Blinova K, Stohlman J, Vicente J, Chan D, Johannesen L, Hortigon-Vinagre MP, Zamora V, Smith G , Crumb WJ, Pang L, Lyn-Cook B. Comprehensive translational assessment of human-induced pluripotent stem cell derived cardiomyocytes for evaluating drug-induced arrhythmias. Toxicological Sciences. 2017 Jan 1;155(1):234-47.
- CiPA Initiative. The Comprehensive In Vitro Proarrhythmia Assay (CiPA) Guide: A New Approach to Cardiac Risk Assessment.
- Ju R, Tian S, Shang Y, Ma S, Zhang M, Liu J, Sun K, Cui L, Zhou X, Han Y. Hepatocyte-like cells and liver organoids: the application of iPSCs and their derivants for treating liver diseases. Materials Advances. 2024;5(21):8419-31.
- Volpato V, Webber C. Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility. Dis. Model. Mech. 13, dmm042317 [Internet]. 2020
- NIH Common Fund. Complement Animal Research In Experimentation (Complement-ARIE). Available at: https://commonfund.nih.gov/complementarie. Accessed 23/09/2025.
.png?width=746&height=390&name=SLAS%20webinar%20%20(1).png)