Application note
Phenotypic characterisation of iPSC-derived ALS disease models by high-throughput MEA
Functional phenotypic characterisation of precision reprogrammed human iPSC-derived disease models for ALS using high-throughput microelectrode arrays In this Application Note, Charles River Laboratories characterised an ALS-relevant phenotype in bit.bio’s ioGlutamatergic Neurons TDP-43 M337V using the Maestro Pro multiwell microelectrode array platform from Axion BioSystems.
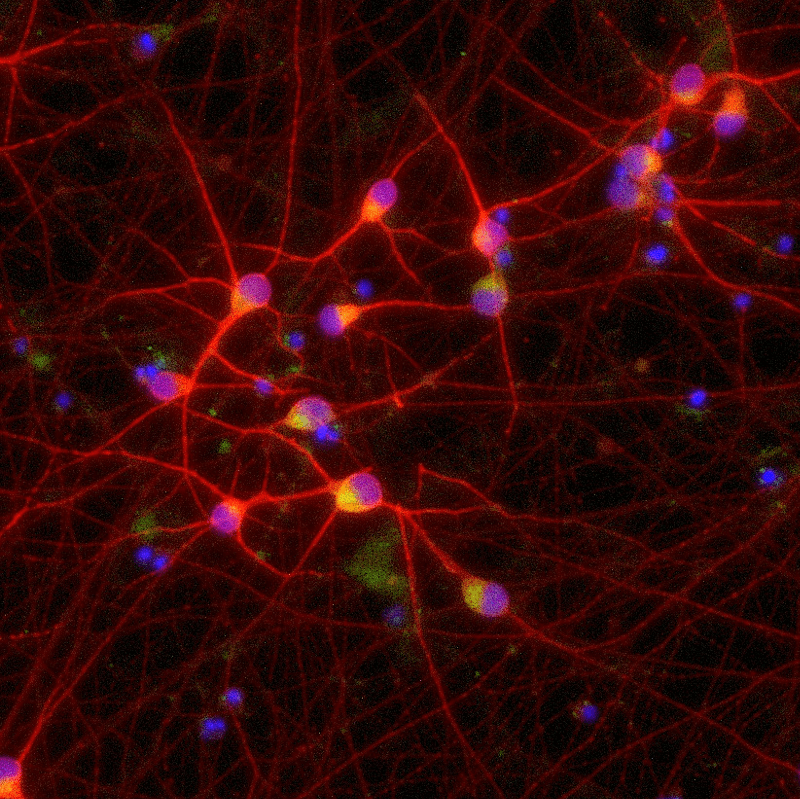
Functional phenotypic characterisation of precision reprogrammed human iPSC-derived disease models for ALS using high-throughput microelectrode arrays In this Application Note, Charles River Laboratories characterised an ALS-relevant phenotype in bit.bio’s ioGlutamatergic Neurons TDP-43 M337V using the Maestro Pro multiwell microelectrode array platform from Axion BioSystems.
In this Application Note, Charles River Laboratories (Cambridge, UK) functionally characterised ioGlutamatergic Neurons TDP-43 M337V/WT, ioGlutamatergic Neurons TDP-43 M337V/M337V and wild-type ioGlutamatergic Neurons developed by bit.bio (Cambridge, UK). Together, these cells form a genetically matched disease model for the investigation of ALS in vitro. For this purpose, they used the Axion Maestro Pro multiwell microelectrode array (MEA) platform to assess electrophysiological differences between the disease model cells and the wild-type control, observing decreased neural activity in the homozygous disease model.
In this application note you will learn:
- How Charles River Laboratories utilised the Maestro Pro multiwell MEA platform from Axion BioSystems to functionally characterise ioGlutamatergic Neurons TDP-43 M337V from bit.bio.
- How ioGluamatergic Neurons TDP-43 M337V and ioGlutamatergic Neurons form a genetically matched model system that allows scientists to attribute experimental outcomes directly to mutations in the TDP-43 gene associated with ALS.
- How using the Axion MEA platform, Charles River generated phenotypic data showing bit.bio's ALS disease models exhibit reduced neuronal activity and network formation compared to a genetically matched control.
V1
2023
bit.bio | Axion BioSystems | Charles River Laboratories